








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (9): 84-96.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0149
Previous Articles Next Articles
CHENG Ya-nan1( ), ZHANG Wen-cong1, ZHOU Yuan2, SUN Xue1, LI Yu1(
), ZHANG Wen-cong1, ZHOU Yuan2, SUN Xue1, LI Yu1( ), LI Qing-gang1(
), LI Qing-gang1( )
)
Received:2023-02-22
Online:2023-09-26
Published:2023-10-24
Contact:
LI Yu, LI Qing-gang
E-mail:379165649@qq.com;liyu@tust.edu.cn;liqinggang@tust.edu.cn
CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium[J]. Biotechnology Bulletin, 2023, 39(9): 84-96.
| 类别 Category | 名称 Name | 特性 Characteristic | 来源 Source |
|---|---|---|---|
| 菌株 Strains | E. coli JM109 | Gene cloning | Lab stock |
| E. coli MG1655 | Give genes manA, manB, gmd, wcaG | Lab stock | |
| L. lactis NZ3900 | A strain of L. lactis | Lab stock | |
| L. lactis NZ9000 | A strain of L. lactis | Lab stock | |
| NZ3900(pNZ8148-1) | Cloning plasmid pNZ8148-1 into NZ3900 | This study | |
| NZ9000(pNZ8148-1) | Cloning plasmid pNZ8148-1 into NZ9000 | This study | |
| NZ3900(pNZ8148-2f) | Cloning plasmid pNZ8148-2f into NZ3900 | This study | |
| NZ9000(pNZ8148-2f) | Cloning plasmid pNZ8148-2f into NZ9000 | This study | |
| NZ9000 1 | Cloning P32, manB, manA, Pnis, gmd, wcaG into NZ9000 | This study | |
| NZ9000 2 | Cloning Pnis, manB, manA, P32, gmd, wcaG into NZ9000 | This study | |
| NZ9000 3 | Cloning P32, manB, manA, gmd, wcaG into NZ9000 | This study | |
| NZ9000 4 | Cloning Pnis, manB, manA, gmd, wcaG into NZ9000 | This study | |
| NZ9000 5 | Cloning plasmid pNZ8148-CfF1 into NZ9000 1 | This study | |
| NZ9000 6 | Cloning plasmid pNZ8148-CfF2 into NZ9000 1 | This study | |
| NZ9000 7 | Cloning plasmid pNZ8148-CfF1 into NZ9000 2 | This study | |
| NZ9000 8 | Cloning plasmid pNZ8148-CfF2 into NZ9000 2 | This study | |
| NZ9000 9 | Cloning plasmid pNZ8148-CfF1 into NZ9000 3 | This study | |
| NZ9000 10 | Cloning plasmid pNZ8148-CfF2 into NZ9000 3 | This study | |
| NZ9000 11 | Cloning plasmid pNZ8148-CfF1 into NZ9000 4 | This study | |
| NZ9000 12 | Cloning plasmid pNZ8148-CfF2 into NZ9000 4 | This study | |
| 质粒 Plasmids | pNZ-fkp | Give gene fkp, GenBank accession NO:CP036555.1 | Lab stock |
| pET-futC | Give gene futC, GenBank accession NO:CP000241.1 | Lab stock | |
| pNZ8148-1 | Cmr, L.lactis expression vector, nisin controlled gene expression(NICE)system, food grade, 2 550 bp | This study | |
| pNZ8148-2f | Cloning fkp, futC and lacF into pNZ8148-1 | This study | |
| pNZ8148-CfF1 | Cloning manC, futC and lacF into pNZ8148-1 | This study | |
| pNZ8148-CfF2 | Cloning manC, P32, futC and lacF into pNZ8148-1 | This study | |
| pNZ5319 | Cmr, GenBank accession NO: DQ104847.1 | Lab stock | |
| pNZ5319-UBAgwD1 | Cloning P32, manB, manA, Pnis, gmd and wcaG into pNZ5319 | This study | |
| pNZ5319-UBAgwD2 | Cloning Pnis, manB, manA, P32, gmd and wcaG into pNZ5319 | This study | |
| pNZ5319-UBAgwD3 | Cloning P32, manB, manA, gmd and wcaG into pNZ5319 | This study | |
| pNZ5319-UBAgwD4 | Cloning Pnis, manB, manA, gmd and wcaG into pNZ5319 | This study | |
| 引物 Primers | 48-F | ataaaagaggagcaaagtaagcactcacccggggtact | |
| 48-R | gactcagcagcttctgcatataatttattttgtagttccttcgaac | ||
| fkp-F1 | atgcagaagctgctgagtct | ||
| fkp-R1 | ttagctacggctaacctgaa | ||
| futC-F1 | ttcaggttagccgtagctaaatggcttttaaagtggtgca | ||
| futC-R1 | gtcatctcttctctgttcacttaagcgttatacttttgggatttt | ||
| lacF-F | gtgaacagagaagagatgactc | ||
| lacF-R | ttactttgctcctcttttataaagttcg | ||
| 5319-F | cgccccatggtagaacttgcactatcaacacactcttaagt | ||
| 5319-R | atgagtcccagaccctccagtatcttaaaattttgtataataggaattg | ||
| up-F | ggagggtctgggactcatcttttc | ||
| up-R | tttgattttcctttcgctagatccctttgc | ||
| down-F | gaaaaattcccttatttttgatatgac | ||
| down-R | aagttctaccatggggcgataac | ||
| manB-F | atgaaaaaattaacctgctttaaag | ||
| manB-R | gcctcctaaatttttatcttactcgttcagcaacgtcag | ||
| manA-F | aatttaggaggcatatcaaatgcaaaaactcattaactcag | ||
| manA-R | ttacagcttgttgtaaacacg | ||
| manA-R1 | atgagagcgacttttgacatttgatatgcctcctaattacagcttgttgtaaacacg | ||
| gmd-F | aaatgtcaaaagtcgctctcat | ||
| gmd-R | gcctcctaaatttttatcttatgactccagcgcgatcg | ||
| wcag-F | aatttaggaggcatatcaaatgagtaaacaacgagtttttattg | ||
| wcag-R | aaaaataagggaatttttcttacccccgaaagcggtcttg | ||
| P32-F1 | ctagcgaaaggaaaatcaaaaattcggtcctcgggatatgataag | ||
| P32-R1 | ctttaaagcaggttaattttttcatatttttatctacctagtata | ||
| P32-F2 | gtgtttacaacaagctgtaaaattcggtcctcgggatatgataag | ||
| P32-R2 | atgagagvgavttttgacatttgatatgcctcctaaatttttatctacctag | ||
| Pnis-F1 | cgtgtttacaacaagctgtaactagtcttataactatactg | ||
| Pnis-R1 | atgagagcgacttttgacatttgatatgcctcctaaataatttattttgtagttcc | ||
| Pnis-F2 | agcgaaaggaaaatcaaatagtcttataactatactgaca | ||
| Pnis-R2 | gcaggttaattttttcatataatttattttgtagttcc | ||
| U-F | gataaaatcaaaacggctaaaatatc | ||
| U-R | gcattaaacgttgaacttttttatggt | ||
| pz-F | cgctacggacgggtgtaaatggcttttaaagtggtgca | ||
| pz-R | gagtttcgactgcgccatataatttattttgtagttccttcg | ||
| manC-F | atggcgcagtcgaaactc | ||
| manC-R | ttacacccgtccgtagcg | ||
| FP-F1 | tactaggtagataaaaatatggcttttaaagtggtgca | ||
| FP-R1 | atcccgaggaccgaatttttacacccgtccgtagcg | ||
| P32-F3 | aaattcggtcctcgggat | ||
| P32-R3 | atttttatctacctagtatagcattt |
Table 1 Strains, plasmids and primers used in this study
| 类别 Category | 名称 Name | 特性 Characteristic | 来源 Source |
|---|---|---|---|
| 菌株 Strains | E. coli JM109 | Gene cloning | Lab stock |
| E. coli MG1655 | Give genes manA, manB, gmd, wcaG | Lab stock | |
| L. lactis NZ3900 | A strain of L. lactis | Lab stock | |
| L. lactis NZ9000 | A strain of L. lactis | Lab stock | |
| NZ3900(pNZ8148-1) | Cloning plasmid pNZ8148-1 into NZ3900 | This study | |
| NZ9000(pNZ8148-1) | Cloning plasmid pNZ8148-1 into NZ9000 | This study | |
| NZ3900(pNZ8148-2f) | Cloning plasmid pNZ8148-2f into NZ3900 | This study | |
| NZ9000(pNZ8148-2f) | Cloning plasmid pNZ8148-2f into NZ9000 | This study | |
| NZ9000 1 | Cloning P32, manB, manA, Pnis, gmd, wcaG into NZ9000 | This study | |
| NZ9000 2 | Cloning Pnis, manB, manA, P32, gmd, wcaG into NZ9000 | This study | |
| NZ9000 3 | Cloning P32, manB, manA, gmd, wcaG into NZ9000 | This study | |
| NZ9000 4 | Cloning Pnis, manB, manA, gmd, wcaG into NZ9000 | This study | |
| NZ9000 5 | Cloning plasmid pNZ8148-CfF1 into NZ9000 1 | This study | |
| NZ9000 6 | Cloning plasmid pNZ8148-CfF2 into NZ9000 1 | This study | |
| NZ9000 7 | Cloning plasmid pNZ8148-CfF1 into NZ9000 2 | This study | |
| NZ9000 8 | Cloning plasmid pNZ8148-CfF2 into NZ9000 2 | This study | |
| NZ9000 9 | Cloning plasmid pNZ8148-CfF1 into NZ9000 3 | This study | |
| NZ9000 10 | Cloning plasmid pNZ8148-CfF2 into NZ9000 3 | This study | |
| NZ9000 11 | Cloning plasmid pNZ8148-CfF1 into NZ9000 4 | This study | |
| NZ9000 12 | Cloning plasmid pNZ8148-CfF2 into NZ9000 4 | This study | |
| 质粒 Plasmids | pNZ-fkp | Give gene fkp, GenBank accession NO:CP036555.1 | Lab stock |
| pET-futC | Give gene futC, GenBank accession NO:CP000241.1 | Lab stock | |
| pNZ8148-1 | Cmr, L.lactis expression vector, nisin controlled gene expression(NICE)system, food grade, 2 550 bp | This study | |
| pNZ8148-2f | Cloning fkp, futC and lacF into pNZ8148-1 | This study | |
| pNZ8148-CfF1 | Cloning manC, futC and lacF into pNZ8148-1 | This study | |
| pNZ8148-CfF2 | Cloning manC, P32, futC and lacF into pNZ8148-1 | This study | |
| pNZ5319 | Cmr, GenBank accession NO: DQ104847.1 | Lab stock | |
| pNZ5319-UBAgwD1 | Cloning P32, manB, manA, Pnis, gmd and wcaG into pNZ5319 | This study | |
| pNZ5319-UBAgwD2 | Cloning Pnis, manB, manA, P32, gmd and wcaG into pNZ5319 | This study | |
| pNZ5319-UBAgwD3 | Cloning P32, manB, manA, gmd and wcaG into pNZ5319 | This study | |
| pNZ5319-UBAgwD4 | Cloning Pnis, manB, manA, gmd and wcaG into pNZ5319 | This study | |
| 引物 Primers | 48-F | ataaaagaggagcaaagtaagcactcacccggggtact | |
| 48-R | gactcagcagcttctgcatataatttattttgtagttccttcgaac | ||
| fkp-F1 | atgcagaagctgctgagtct | ||
| fkp-R1 | ttagctacggctaacctgaa | ||
| futC-F1 | ttcaggttagccgtagctaaatggcttttaaagtggtgca | ||
| futC-R1 | gtcatctcttctctgttcacttaagcgttatacttttgggatttt | ||
| lacF-F | gtgaacagagaagagatgactc | ||
| lacF-R | ttactttgctcctcttttataaagttcg | ||
| 5319-F | cgccccatggtagaacttgcactatcaacacactcttaagt | ||
| 5319-R | atgagtcccagaccctccagtatcttaaaattttgtataataggaattg | ||
| up-F | ggagggtctgggactcatcttttc | ||
| up-R | tttgattttcctttcgctagatccctttgc | ||
| down-F | gaaaaattcccttatttttgatatgac | ||
| down-R | aagttctaccatggggcgataac | ||
| manB-F | atgaaaaaattaacctgctttaaag | ||
| manB-R | gcctcctaaatttttatcttactcgttcagcaacgtcag | ||
| manA-F | aatttaggaggcatatcaaatgcaaaaactcattaactcag | ||
| manA-R | ttacagcttgttgtaaacacg | ||
| manA-R1 | atgagagcgacttttgacatttgatatgcctcctaattacagcttgttgtaaacacg | ||
| gmd-F | aaatgtcaaaagtcgctctcat | ||
| gmd-R | gcctcctaaatttttatcttatgactccagcgcgatcg | ||
| wcag-F | aatttaggaggcatatcaaatgagtaaacaacgagtttttattg | ||
| wcag-R | aaaaataagggaatttttcttacccccgaaagcggtcttg | ||
| P32-F1 | ctagcgaaaggaaaatcaaaaattcggtcctcgggatatgataag | ||
| P32-R1 | ctttaaagcaggttaattttttcatatttttatctacctagtata | ||
| P32-F2 | gtgtttacaacaagctgtaaaattcggtcctcgggatatgataag | ||
| P32-R2 | atgagagvgavttttgacatttgatatgcctcctaaatttttatctacctag | ||
| Pnis-F1 | cgtgtttacaacaagctgtaactagtcttataactatactg | ||
| Pnis-R1 | atgagagcgacttttgacatttgatatgcctcctaaataatttattttgtagttcc | ||
| Pnis-F2 | agcgaaaggaaaatcaaatagtcttataactatactgaca | ||
| Pnis-R2 | gcaggttaattttttcatataatttattttgtagttcc | ||
| U-F | gataaaatcaaaacggctaaaatatc | ||
| U-R | gcattaaacgttgaacttttttatggt | ||
| pz-F | cgctacggacgggtgtaaatggcttttaaagtggtgca | ||
| pz-R | gagtttcgactgcgccatataatttattttgtagttccttcg | ||
| manC-F | atggcgcagtcgaaactc | ||
| manC-R | ttacacccgtccgtagcg | ||
| FP-F1 | tactaggtagataaaaatatggcttttaaagtggtgca | ||
| FP-R1 | atcccgaggaccgaatttttacacccgtccgtagcg | ||
| P32-F3 | aaattcggtcctcgggat | ||
| P32-R3 | atttttatctacctagtatagcattt |

Fig. 2 Construction of plasmids required for the de novo synthesis of 2'-FL A: Construction process of the plasmids expressing pathway genes. B: Construction process of plasmids for gene recombination. The number indicates the construction steps of each fragment and plasmid
| 胰蛋白胨 Tryptone/(g·L-1) | 酵母粉 Yeast extract/(g·L-1) | |||
|---|---|---|---|---|
| 5 | 6 | 8 | 10 | 12 |
| 10 | 5 | 7 | 9 | 11 |
| 15 | 4 | 6 | 8 | 10 |
| 20 | 3 | 5 | 7 | 9 |
Table 2 Co-optimized concentration combination of tryptone and yeast extract
| 胰蛋白胨 Tryptone/(g·L-1) | 酵母粉 Yeast extract/(g·L-1) | |||
|---|---|---|---|---|
| 5 | 6 | 8 | 10 | 12 |
| 10 | 5 | 7 | 9 | 11 |
| 15 | 4 | 6 | 8 | 10 |
| 20 | 3 | 5 | 7 | 9 |
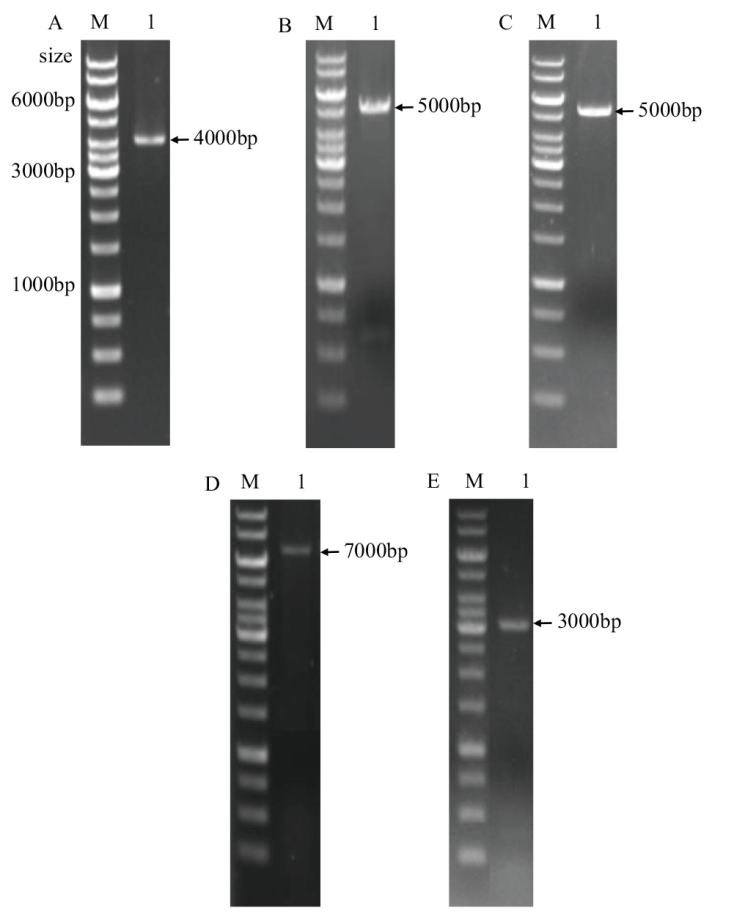
Fig. 3 Electrophoresis diagram of PCR product for strain validation A: Verification of plasmids pNZ8148-2f. B: Verification of plasmid pNZ5319-UBAgwD1. C: Strain NZ9000 1 was verified by single crossover. D: Strain NZ9000 1 was verified by double crossover. E: Verification of plasmid pNZ8148-CfF1. M: Trans 1 K DNA marker. The sizes of the main bands were signed
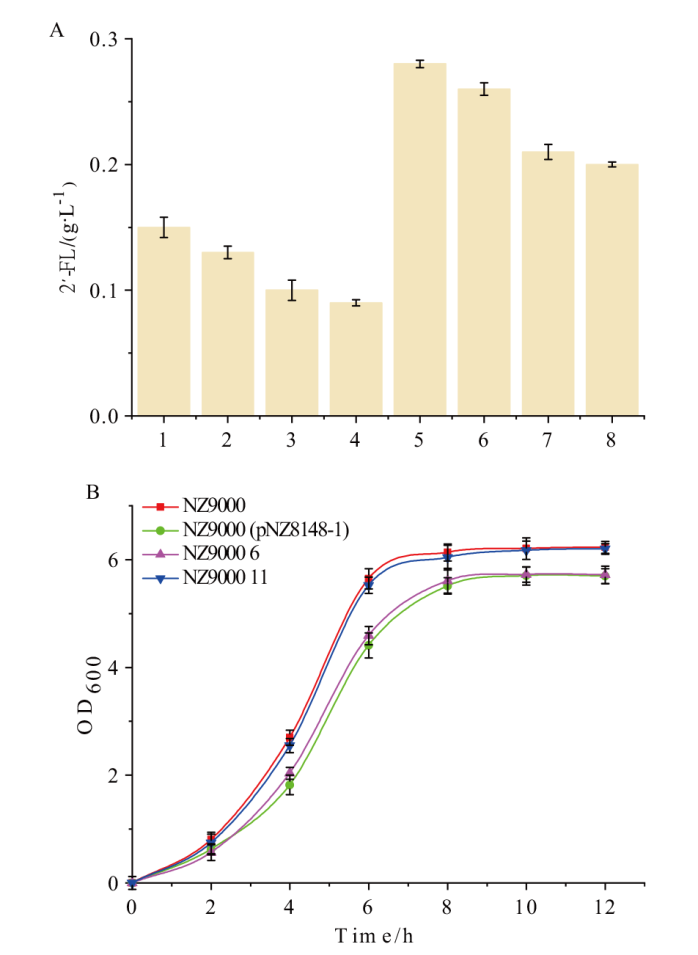
Fig. 5 Productions and growth curves of different strains with 2'-FL de novo synthesis pathway A: The 2'-FL production of different strains after 48 h fermentation. 1: NZ9000 5;2: NZ9000 7; 3: NZ9000 9; 4: NZ9000 11; 5: NZ9000 6; 6: NZ9000 8;7: NZ9000 10; 8: NZ9000 12. B: The growth curve of different strains
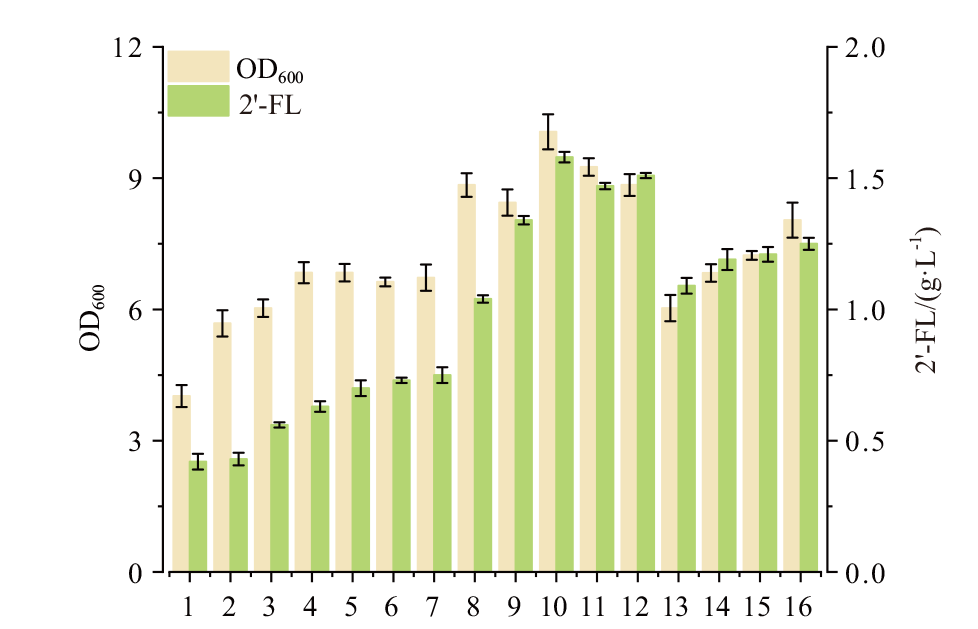
Fig. 7 Effects of tryptone and yeast extract concentration on cell growth and 2'-FL production Number 1-16 represent the cultures with different tryptone and yeast concentration. 1-4: Tryptone set as 5 g/L, yeast extract varied: 6, 8, 10 and 12 g/L. 5-8: Tryptone set as 10 g/L, yeast extract varied: 5, 7, 9 and 11 g/L. 9-12: Tryptone set as 15 g/L, yeast extract varied: 4, 6, 8 and 10 g/L. 13-16: Tryptone set as 20 g/L, yeast extract varied: 3, 5, 7 and 9 g/L
| [1] | 揭良, 苏米亚, 贾宏信, 等. 母乳寡糖的研究进展[J]. 食品工业, 2020, 41(10): 258-261. |
| Jie L, Su MY, Jia HX, et al. Recent progress in research of human milk oligosaccharides[J]. Food Ind, 2020, 41(10): 258-261. | |
| [2] | 陈坚, 邓洁莹, 李江华, 等. 母乳寡糖的生物合成研究进展[J]. 中国食品学报, 2016, 16(11): 1-8. |
| Chen J, Deng JY, Li JH, et al. Advances in biosynthesis of breast milk oligosaccharides[J]. J Chin Inst Food Sci Technol, 2016, 16(11): 1-8. | |
| [3] |
Huang D, Yang KX, Liu J, et al. Metabolic engineering of Escherichia coli for the production of 2'-fucosyllactose and 3-fucosyllactose through modular pathway enhancement[J]. Metab Eng, 2017, 41: 23-38.
doi: S1096-7176(16)30233-6 pmid: 28286292 |
| [4] |
Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens[J]. Annu Rev Nutr, 2005, 25: 37-58.
pmid: 16011458 |
| [5] |
Berger PK, Plows JF, Jones RB, et al. Human milk oligosaccharide 2'-fucosyllactose links feedings at 1 month to cognitive development at 24 months in infants of normal and overweight mothers[J]. PLoS One, 2020, 15(2): e0228323.
doi: 10.1371/journal.pone.0228323 URL |
| [6] |
Matthies H, Staak S, Krug M. Fucose and fucosyllactose enhance in-vitro hippocampal long-term potentiation[J]. Brain Res, 1996, 725(2): 276-280.
pmid: 8836537 |
| [7] |
Bienenstock J, Buck RH, Linke H, et al. Fucosylated but not sialylated milk oligosaccharides diminish colon motor contractions[J]. PLoS One, 2013, 8(10): e76236.
doi: 10.1371/journal.pone.0076236 URL |
| [8] |
Petschacher B, Nidetzky B. Biotechnological production of fucosylated human milk oligosaccharides: Prokaryotic fucosyltransferases and their use in biocatalytic cascades or whole cell conversion systems[J]. J Biotechnol, 2016, 235: 61-83.
doi: 10.1016/j.jbiotec.2016.03.052 pmid: 27046065 |
| [9] |
Wang B. Sialic acid is an essential nutrient for brain development and cognition[J]. Annu Rev Nutr, 2009, 29: 177-222.
doi: 10.1146/annurev.nutr.28.061807.155515 pmid: 19575597 |
| [10] |
Vazquez E, Barranco A, Ramirez M, et al. Dietary 2'-fucosyllactose enhances operant conditioning and long-term potentiation via gut-brain communication through the vagus nerve in rodents[J]. PLoS One, 2016, 11(11): e0166070.
doi: 10.1371/journal.pone.0166070 URL |
| [11] |
Albermann C, Distler J, Piepersberg W. Preparative synthesis of GDP-β-L-fucose by recombinant enzymes from enterobacterial sources[J]. Glycobiology, 2000, 10(9): 875-881.
pmid: 10988249 |
| [12] |
Becker DJ, Lowe JB. Fucose: biosynthesis and biological function in mammals[J]. Glycobiology, 2003, 13(7): 41R-53R.
pmid: 12651883 |
| [13] |
Coyne MJ, Reinap B, Lee MM, et al. Human symbionts use a host-like pathway for surface fucosylation[J]. Science, 2005, 307(5716): 1778-1781.
pmid: 15774760 |
| [14] |
Liu TW, Ito H, Chiba Y, et al. Functional expression of l-fucokinase/guanosine 5'-diphosphate-l-fucose pyrophosphorylase from Bacteroides fragilis in Saccharomyces cerevisiae for the production of nucleotide sugars from exogenous monosaccharides[J]. Glycobiology, 2011, 21(9): 1228-1236.
doi: 10.1093/glycob/cwr057 URL |
| [15] |
Mattila P, Räbinä J, Hortling S, et al. Functional expression of Escherichia coli enzymes synthesizing GDP-L-fucose from inherent GDP-D-mannose in Saccharomyces cerevisiae[J]. Glycobiology, 2000, 10(10): 1041-1047.
pmid: 11030750 |
| [16] |
Järvinen N, Mäki M, Räbinä J, et al. Cloning and expression of Helicobacter pylori GDP-l-fucose synthesizing enzymes(GMD and GMER)in Saccharomyces cerevisiae[J]. Eur J Biochem, 2001, 268(24): 6458-6464.
pmid: 11737200 |
| [17] |
Liu JJ, Kwak S, Pathanibul P, et al. Biosynthesis of a functional human milk oligosaccharide, 2'-fucosyllactose, and l-fucose using engineered Saccharomyces cerevisiae[J]. ACS Synth Biol, 2018, 7(11): 2529-2536.
doi: 10.1021/acssynbio.8b00134 URL |
| [18] |
Yu S, Liu JJ, Yun EJ, et al. Production of a human milk oligosaccharide 2'-fucosyllactose by metabolically engineered Saccharomyces cerevisiae[J]. Microb Cell Fact, 2018, 17(1): 101.
doi: 10.1186/s12934-018-0947-2 |
| [19] |
Chin YW, Park JB, Park YC, et al. Metabolic engineering of Corynebacterium glutamicum to produce GDP-l-fucose from glucose and mannose[J]. Bioprocess Biosyst Eng, 2013, 36(6): 749-756.
doi: 10.1007/s00449-013-0900-z URL |
| [20] | Shin CS, Yoon JW, Song YH, et al. Method of producing 2'-fucosyllactose using Corynebacterium glutamicum: US20180298389[P]. 2018-10-18. |
| [21] |
Song AAL, In LLA, Lim SHE, et al. A review on Lactococcus lactis: from food to factory[J]. Microb Cell Fact, 2017, 16(1): 55.
doi: 10.1186/s12934-017-0669-x URL |
| [22] |
García-Fruitós E. Lactic acid bacteria: a promising alternative for recombinant protein production[J]. Microb Cell Fact, 2012, 11(1): 157.
doi: 10.1186/1475-2859-11-157 URL |
| [23] | 尹建洪, 罗立新, 王成. 南极假丝酵母脂肪酶B(CALB)基因在乳酸乳球菌MG1363中的整合及表达[J]. 生物技术通报, 2012(5): 105-109. |
| Yin JH, Luo LX, Wang C. Expression and integration of Candida antarctica lipase B gene in Lactococcus lactis MG1363[J]. Biotechnol Bull, 2012(5): 105-109. | |
| [24] |
Villena J, Medina M, Racedo S, et al. Resistance of young mice to pneumococcal infection can be improved by oral vaccination with recombinant Lactococcus lactis[J]. J Microbiol Immunol Infect, 2010, 43(1): 1-10.
doi: 10.1016/S1684-1182(10)60001-1 pmid: 20434117 |
| [25] |
Lokman BC, Heerikhuisen M, Leer RJ, et al. Regulation of expression of the Lactobacillus pentosus xylAB operon[J]. J Bacteriol, 1997, 179(17): 5391-5397.
pmid: 9286992 |
| [26] |
Ferain T, Hobbs JN Jr, Richardson J, et al. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum[J]. J Bacteriol, 1996, 178(18): 5431-5437.
pmid: 8808932 |
| [27] |
Sun X, Peng ZT, Li C, et al. Combinatorial metabolic engineering and tolerance evolving of Escherichia coli for high production of 2'-fucosyllactose[J]. Bioresour Technol, 2023, 372: 128667.
doi: 10.1016/j.biortech.2023.128667 URL |
| [28] |
Gioia MG, Andreatta P, Boschetti S, et al. Development and validation of a liquid chromatographic method for the determination of branched-chain amino acids in new dosage forms[J]. J Pharm Biomed Anal, 2007, 45(3): 456-464.
pmid: 17875379 |
| [29] | 瓮茹茹, 卫鑫慧, 李浩正, 等. 2'-岩藻糖基乳糖的微生物合成研究进展[J]. 食品科学, 2021, 42(17): 248-254. |
| Weng RR, Wei XH, Li HZ, et al. Progress in microbial synthesis of 2'-fucosyllactose[J]. Food Sci, 2021, 42(17): 248-254. | |
| [30] | 徐铮, 李娜, 陈盈利, 等. 人乳寡糖2'-FL和3-FL的生物制备研究进展[J]. 生物工程学报, 2020, 36(12): 2767-2778. |
| Xu Z, Li N, Chen YL, et al. Recent advances in the bio-production of human milk oligosaccharides 2'-FL and 3-FL[J]. Chin J Biotechnol, 2020, 36(12): 2767-2778. | |
| [31] |
Baumgärtner F, Seitz L, Sprenger GA, et al. Construction of Escherichia coli strains with chromosomally integrated expression cassettes for the synthesis of 2'-fucosyllactose[J]. Microb Cell Fact, 2013, 12: 40.
doi: 10.1186/1475-2859-12-40 pmid: 23635327 |
| [32] | 贾红红, 李玉, 刘逸寒, 等. α1, 2-岩藻糖基转移酶基因的克隆及在大肠杆菌中的表达[J]. 生物技术通报, 2011(3): 185-190. |
| Jia HH, Li Y, Liu YH, et al. Cloning and expression of alpha 1, 2-fucosyltransferase in E. coli[J]. Biotechnol Bull, 2011(3): 185-190. | |
| [33] |
白凤翎, 张柏林, 蒋湘宁. 乳酸菌有氧呼吸代谢研究进展[J]. 食品科学, 2009, 30(13): 262-267.
doi: 10.7506/spkx1002-6630-200913061 |
|
Bai FL, Zhang BL, Jiang XN. Research progress in aerobic respiration metabolism of lactic acid bacteria[J]. Food Sci, 2009, 30(13): 262-267.
doi: 10.1111/jfds.1965.30.issue-2 URL |
|
| [34] | 付龙云. 乳酸菌抗氧胁迫及有氧生长的研究[D]. 济南: 山东大学, 2013. |
| Fu LY. Study on anti-oxygen stress and aerobic growth of lactic acid bacteria[D]. Jinan: Shandong University, 2013. | |
| [35] |
Deng JY, Gu LY, Chen TC, et al. Engineering the substrate transport and cofactor regeneration systems for enhancing 2'-fucosyllactose synthesis in Bacillus subtilis[J]. ACS Synth Biol, 2019, 8(10): 2418-2427.
doi: 10.1021/acssynbio.9b00314 URL |
| [1] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [2] | ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering [J]. Biotechnology Bulletin, 2023, 39(8): 137-147. |
| [3] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [4] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [5] | YU Hui-li, LI Ai-tao. Application of Cytochrome P450 in the Biosynthesis of Flavors and Fragrances [J]. Biotechnology Bulletin, 2023, 39(4): 24-37. |
| [6] | QIU Yi-bin, MA Yan-qin, SHA Yuan-yuan, ZHU Yi-fan, SU Er-zheng, LEI Peng, LI Sha, XU Hong. Research Progress in Molecular Genetic Manipulation Technology of Bacillus amyloliquefaciens and Its Application [J]. Biotechnology Bulletin, 2022, 38(2): 205-217. |
| [7] | MA Yan-qin, QIU Yi-bin, LI Sha, XU Hong. Research Progress in the Biosynthesis and Metabolic Engineering of Hyaluronic Acid [J]. Biotechnology Bulletin, 2022, 38(2): 252-262. |
| [8] | ZHOU Jing, HUANG Wen-mao, QIN Li-jun, HAN Li-zhen. Construction of Mixed Fermentation System of Four PGPR Strains and Evaluation of Its Promoting Effect [J]. Biotechnology Bulletin, 2021, 37(4): 116-126. |
| [9] | ZHANG Yao-xin, WANG Liang-jie, ZHENG Wen, XU Han-qin, ZHENG Lian, ZHONG Jing. Study on Enzyme Production of a Chitinase-producing Strain Achromobacter sp. ZWW8 by Fermentation and Its Enzymatic Characterization [J]. Biotechnology Bulletin, 2021, 37(4): 96-106. |
| [10] | WU Rong, CAO Jia-rui, CAO Jun, LIU Fei-xiang, YANG Meng, SU Er-zheng. Expression and Fermentation Optimization of Candida antarctica Lipase B in Escherichia coli [J]. Biotechnology Bulletin, 2021, 37(2): 138-148. |
| [11] | YE Jian-wen, CHEN Jiang-nan, ZHANG Xu, Wu Fu-qing, CHEN Guo-qiang. Dynamic Control:An Efficient Strategy for Metabolically Engineering Microbial Cell Factories [J]. Biotechnology Bulletin, 2020, 36(6): 1-12. |
| [12] | LI Xiao-yan, LI Ze-qi, WANG Yu-qian, YU Jing, LIN Zhen-ping, LIN Xiang-min. Construction of Aeromonas hydrophila acrA Deficient Strain and Determination of Its Physiological Function [J]. Biotechnology Bulletin, 2020, 36(11): 63-69. |
| [13] | WANG Ke-wen ,YIN Xue, WANG Yu ,LI Yu-hua. Application of Selection and Optimization of Promoter in Metabolic Engineering of Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2018, 34(6): 38-47. |
| [14] | HUANG Fang, LIN Xiang-min. Construction of Mutant Strain bamA,bamB and bamD of Aeromonas hydrophila and Their Effects on the Outer Membrane Protein Transportation [J]. Biotechnology Bulletin, 2018, 34(5): 148-153. |
| [15] | LIN Bei, LI Jian-Xiu, LIU Xue-ling. The Effects of By-products of Hydrolyzing Lignocellulose on Ethanol Fermentation and Relevant Countermeasures [J]. Biotechnology Bulletin, 2018, 34(3): 23-30. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||