








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (12): 113-123.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0325
Previous Articles Next Articles
SUN Dan-ni( ), LI Hao, CUI Yu-meng, HUANG He(
), LI Hao, CUI Yu-meng, HUANG He( )
)
Received:2024-04-06
Online:2024-12-26
Published:2025-01-15
Contact:
HUANG He
E-mail:sundanni@bjfu.edu.cn;101navy@163.com
SUN Dan-ni, LI Hao, CUI Yu-meng, HUANG He. Study on the Regulation of Anthocyanin Metabolism by miR156-PhSPL3 in Pericallis hybrida[J]. Biotechnology Bulletin, 2024, 40(12): 113-123.
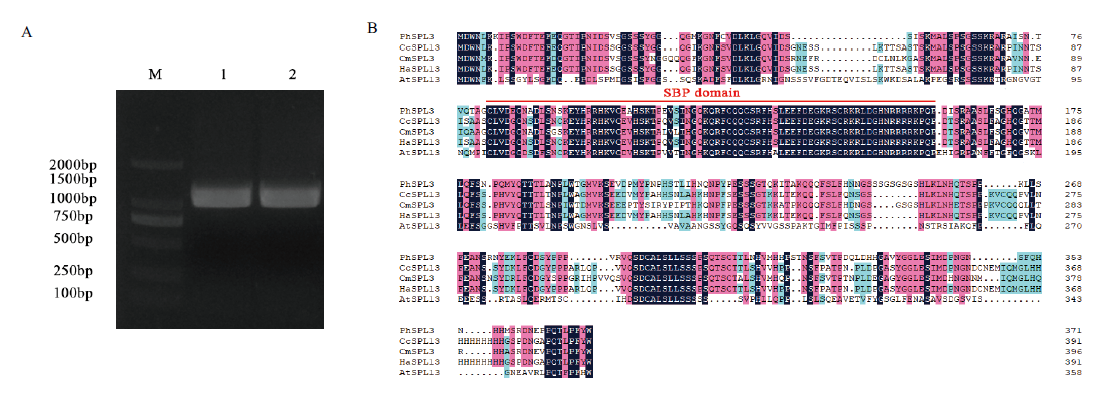
Fig. 1 Cloning of PhSPL3 gene and alignment of SPL amino acid sequences with those from other species A: Cloning of PhSPL3 gene; M: DL2000 DNA Marker; 1: amplification using cDNA from PeC ray florets as template; 2: amplification using cDNA from PeB ray florets as template. B: Amino acid sequence alignment of PhSPL3 protein; PhSPL3: Pericallis hybrida(Unigene0015241); CcSPL13: Cynara cardunculus var. scolymus(XP_024977060.1); CmSPL3: Chrysanthemum ×morifolium(ALF46633.1); HaSPL3: Helianthus annuus(XP_021978557.1); AtSPL13: Arabidopsis thaliana(AT5G50570). The red lines in the figure B indicate the positions of the SBP domain

Fig. 2 Cloning and sequence alignment of Ph-miR156 precursor genes A: Cloning of Ph-miR156 precursor gene; M: DL2000 DNA Marker; 1: cloning of Ph-MIR156a gene; 2: cloning of Ph-MIR156b gene. B: Sequence alignment of Ph-MIR156a and Ph-MIR156b. The red lines in the figure B indicate mature miR156 sequences
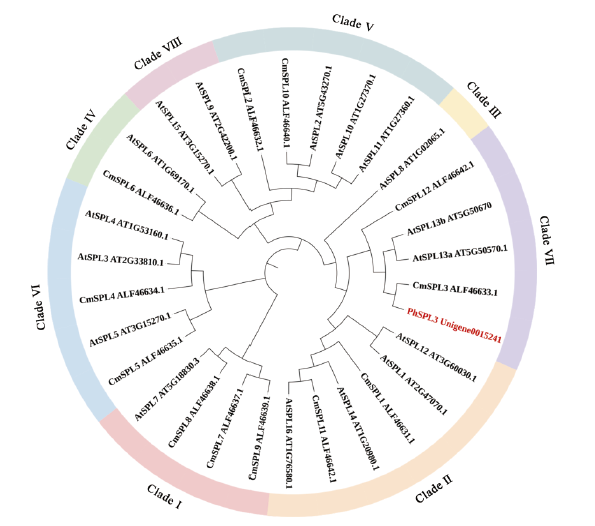
Fig. 3 Phylogenetic analysis of PhSPL3 protein family in comparison with SPL protein from A. thaliana and C. ×morifolium At: Arabidopsis thaliana; Cm: Chrysanthemum ×morifolium; Ph: Pericallis hybrida
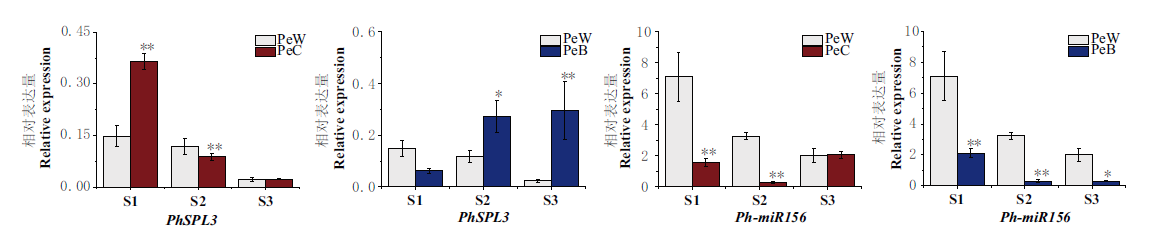
Fig. 4 RT-qPCR expression analysis of PhSPL3 and Ph-miR156 in ray florets of PeW, PeC and PeB at stage S1-S3 PeW: White Pericallis hybrida, PeC: Carmine Pericallis hybrida, PeB: Blue Pericallis hybrida. * and ** indicate significant differences at 0.05 and 0.01 levels
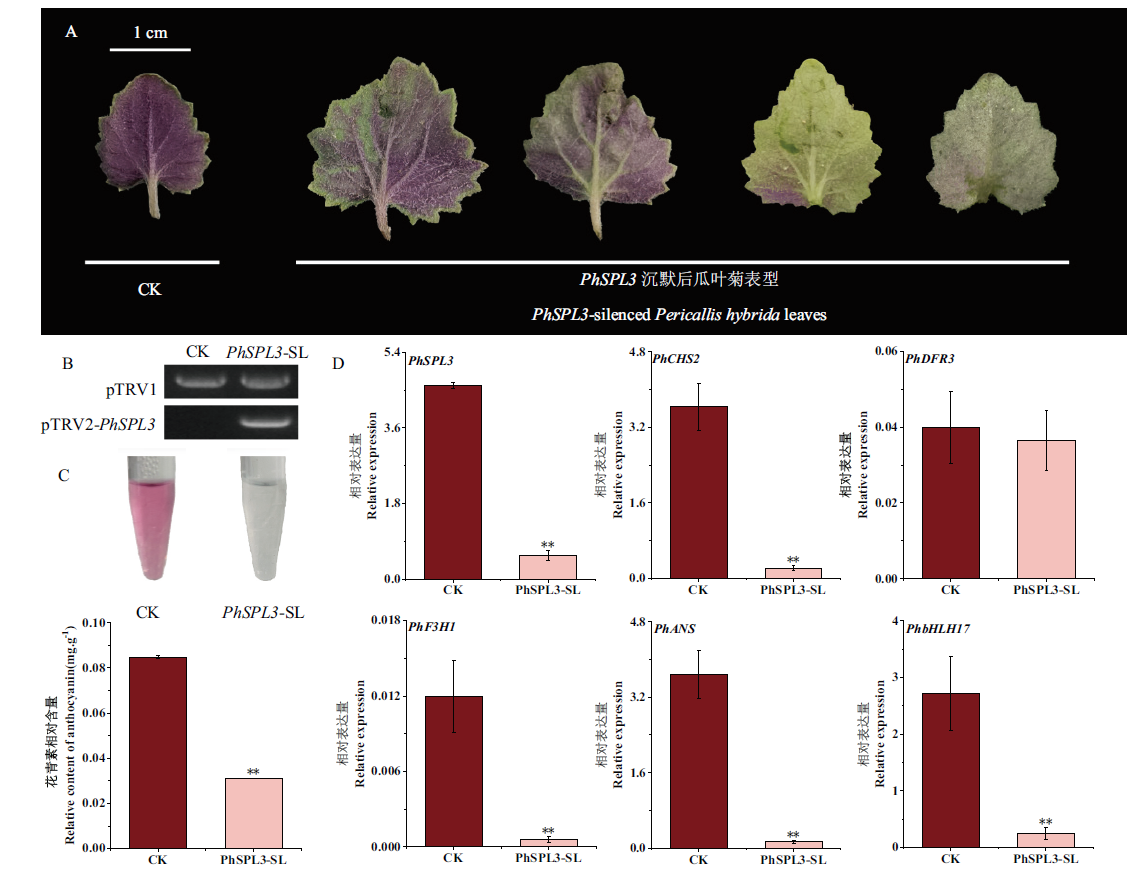
Fig. 6 Transient silencing of PhSPL3 by VIGS in the leaves of PeC A: Phenotypes of CK and PhPSL3-silenced Pericallis hybrida leaves. B:Detection of pTRV1 and pTRV2-PhSPL3 in the silenced tissue. C: Quantitative analysis of anthocyanins in PhSPL3-silenced leaves and the control. D: Expressions of genes in the anthocyanin biosynthesis pathway in PhSPL3-silenced and the control. CK: Leaf samples infiltrated with pTRV1 and pTRV2::00. PhSPL3-SL: Leaf samples infiltrated with pTRV1 and pTRV2::PhSPL3. * and ** indicate significant differences at 0.05 and 0.01 levels respectively
| [1] | 赵启明, 李范, 李萍. 花青素生物合成关键酶的研究进展[J]. 生物技术通报, 2012(12): 25-32. |
| Zhao QM, Li F, Li P. Research advances on core enzymes of anthocyanidin biosynthesis[J]. Biotechnol Bull, 2012(12): 25-32. | |
| [2] | Gould K, Davies KM, Winefield C. Anthocyanins: biosynthesis, functions, and applications[M]. New York: Springer, 2009. |
| [3] |
刘恺媛, 王茂良, 辛海波, 等. 植物花青素合成与调控研究进展[J]. 中国农学通报, 2021, 37(14): 41-51.
doi: 10.11924/j.issn.1000-6850.casb2020-0390 |
|
Liu KY, Wang ML, Xin HB, et al. Anthocyanin biosynthesis and regulate mechanisms in plants: a review[J]. Chin Agric Sci Bull, 2021, 37(14): 41-51.
doi: 10.11924/j.issn.1000-6850.casb2020-0390 |
|
| [4] |
曾鑫海, 陈锐, 师宇, 等. 植物SPL转录因子的生物功能研究进展[J]. 植物学报, 2023, 58(6):982-997.
doi: 10.11983/CBB22216 |
| Zeng XH, Chen R, Shi Y, et al. Research advances in biological functions of plant SPL transcription factors[J]. Chinese Bulletin of Botany, 2023, 58(6):982-997. | |
| [5] | Gou JY, Felippes FF, Liu CJ, et al. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor[J]. Plant Cell, 2011, 23(4): 1512-1522. |
| [6] | Su ZW, Wang XC, Xuan XX, et al. Characterization and action mechanism analysis of VvmiR156b/c/d-VvSPL9 module responding to multiple-hormone signals in the modulation of grape berry color formation[J]. Foods, 2021, 10(4): 896. |
| [7] | Liu HN, Shu Q, Kui LW, et al. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear[J]. Mol Hortic, 2021, 1(1): 14. |
| [8] | 王璐. 瓜叶菊花青素苷合成分支途径的调控机制[D]. 北京: 北京林业大学, 2015. |
| Wang L. Regulation mechanism of anthocyanin synthesis branching pathway in chrysanthemum morifolium[D]. Beijing: Beijing Forestry University, 2015. | |
| [9] | 金雪花. 基于高通量测序的瓜叶菊花青素苷合成途径研究[D]. 北京: 北京林业大学, 2013. |
| Jin XH. Study on the synthetic pathway of anthocyanin glycosides from Chrysanthemum morifolium based on high-throughput sequencing[D]. Beijing: Beijing Forestry University, 2013. | |
| [10] | 李亚军. 蓝色瓜叶菊聚酰化花青素生物合成相关基因分离和功能分析[D]. 北京: 北京林业大学, 2020. |
| Li YJ. Isolation and functional analysis of genes related to biosynthesis of polyacylanthocyanidins from Chrysanthemum morifolium[D]. Beijing: Beijing Forestry University, 2020. | |
| [11] | Li YJ, Liu YT, Qi FT, et al. Establishment of virus-induced gene silencing system and functional analysis of ScbHLH17 in Senecio cruentus[J]. Plant Physiol Biochem, 2020, 147: 272-279. |
| [12] | Cui YM, Fan JW, Lu CF, et al. ScGST3 and multiple R2R3-MYB transcription factors function in anthocyanin accumulation in Senecio cruentus[J]. Plant Sci, 2021, 313: 111094. |
| [13] | Cui YM, Fan JW, Liu FY, et al. R2R3-MYB transcription factor PhMYB2 positively regulates anthocyanin biosynthesis in Pericallis hybrida[J]. Sci Hortic, 2023, 322: 112446. |
| [14] | Li H, Qi FT, Sun DN, et al. Integrated transcriptome and small RNA sequencing revealing miRNA-mediated regulatory network of bicolour pattern formation in Pericallis hybrida ray florets[J]. Sci Hortic, 2024, 326: 112765. |
| [15] | Qi FT, Liu YT, Luo YL, et al. Functional analysis of the ScAG and ScAGL11 MADS-box transcription factors for anthocyanin biosynthesis and bicolour pattern formation in Senecio cruentus ray florets[J]. Hortic Res, 2022, 9: uhac071. |
| [16] | Lu CF, Li YJ, Cui YM, et al. Isolation and functional analysis of genes involved in polyacylated anthocyanin biosynthesis in blue Senecio cruentus[J]. Front Plant Sci, 2021, 12: 640746. |
| [17] |
Cardon G, Höhmann S, Klein J, et al. Molecular characterisation of the Arabidopsis SBP-box genes[J]. Gene, 1999, 237(1): 91-104.
doi: 10.1016/s0378-1119(99)00308-x pmid: 10524240 |
| [18] | Song AP, Gao TW, Wu D, et al. Transcriptome-wide identification and expression analysis of chrysanthemum SBP-like transcription factors[J]. Plant Physiol Biochem, 2016, 102: 10-16. |
| [19] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔt Method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [20] | 陈文文, 吴怀通, 陈赢男. SPL家族基因复制及功能分化分析[J]. 南京林业大学学报: 自然科学版, 2020, 44(5): 55-66. |
| Chen WW, Wu HT, Chen YN. Gene duplications and functional divergence analyses of the SPL gene family[J]. J Nanjing For Univ Nat Sci Ed, 2020, 44(5): 55-66. | |
| [21] |
Birkenbihl RP, Jach G, Saedler H, et al. Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains[J]. J Mol Biol, 2005, 352(3): 585-596.
doi: 10.1016/j.jmb.2005.07.013 pmid: 16095614 |
| [22] | Xu ML, Hu TQ, Zhao JF, et al. Developmental functions of mir156-regulated squamosa promoter binding protein-like(spl)genes in Arabidopsis thaliana[J]. PLoS Genet, 2016, 12(8): e1006263. |
| [23] | Martin RC, Asahina M, Liu PP, et al. The regulation of post-germinative transition from the cotyledon- to vegetative-leaf stages by microRNA-targeted SQUAMOSA PROMOTER-BINDING PROTEIN LIKE13 in Arabidopsis[J]. Seed Sci Res, 2010, 20(2): 89-96. |
| [24] |
Hu TQ, Manuela D, Xu ML. Squamosa promoter binding protein-like 9 and 13 repress blade-on-petiole 1 and 2 directly to promote adult leaf morphology in Arabidopsis[J]. J Exp Bot, 2023, 74(6): 1926-1939.
doi: 10.1093/jxb/erad017 pmid: 36629519 |
| [25] | Feyissa BA, Arshad M, Gruber MY, et al. The interplay between miR156/SPL13 and DFR/WD40-1 regulate drought tolerance in alfalfa[J]. BMC Plant Biol, 2019, 19(1): 434. |
| [26] |
Yang T, Li KT, Hao SX, et al. The use of RNA sequencing and correlation network analysis to study potential regulators of crabapple leaf color transformation[J]. Plant Cell Physiol, 2018, 59(5): 1027-1042.
doi: 10.1093/pcp/pcy044 pmid: 29474693 |
| [27] | Yu N, Cai WJ, Wang SC, et al. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana[J]. Plant Cell, 2010, 22(7): 2322-2335. |
| [28] | 魏梦苒. 芍药花色形成相关miRNA的挖掘及功能分析研究[D]. 扬州: 扬州大学, 2017. |
| Wei MR. Mining and functional analysis of miRNA related to flower color formation in Paeonia lactiflora[D]. Yangzhou: Yangzhou University, 2017. | |
| [29] | Li HS, Ma B, Luo YW, et al. The mulberry SPL gene family and the response of MnSPL7 to silkworm herbivory through activating the transcription of MnTT2L2 in the catechin biosynthesis pathway[J]. Int J Mol Sci, 2022, 23(3): 1141. |
| [30] | Guo SH, Zhang M, Feng MX, et al. miR156b-targeted VvSBP8/13 functions downstream of the abscisic acid signal to regulate anthocyanins biosynthesis in grapevine fruit under drought[J]. Hortic Res, 2024, 11(2): uhad293. |
| [31] |
Yang T, Ma HY, Zhang J, et al. Systematic identification of long noncoding RNAs expressed during light-induced anthocyanin accumulation in apple fruit[J]. Plant J, 2019, 100(3): 572-590.
doi: 10.1111/tpj.14470 |
| [1] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||