








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (7): 186-193.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1072
Previous Articles Next Articles
YU Qiu-lin1( ), MA Jing-yi1, ZHAO Pan1, SUN Peng-fang1, HE Yu-mei1, LIU Shi-biao2, GUO Hui-hong1(
), MA Jing-yi1, ZHAO Pan1, SUN Peng-fang1, HE Yu-mei1, LIU Shi-biao2, GUO Hui-hong1( )
)
Received:2021-08-20
Online:2022-07-26
Published:2022-08-09
Contact:
GUO Hui-hong
E-mail:YuQiulin@163.com;guohh@bjfu.edu.cn
YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b[J]. Biotechnology Bulletin, 2022, 38(7): 186-193.
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| GpmiR156a-F | TCTTYACWCAACWTCTCACCCA |
| GpmiR156a-R | GAGARAYCWGCATAWCTCAATMACC |
| GpmiR166b-F | CAGATACATGTACGAGGWGTTCA |
| GpmiR166b-R | GCTTCATCATYAYACCAATCTGC |
| GpmiR156a-Ft | TGGAGAGAACACGGGGGACTCTAGA TCTTYACWCAACWTCTCACCCA |
| GpmiR156a-Rt | TAACATAAGGGACTGACCACCCGGG GAGARAYCWGCATAWCTCAATMACC |
| GpmiR166b-Ft | TGGAGAGAACACGGGGGACTCTAGA CAGATA- CATGTACGAGGWGTTCA |
| GpmiR166b-Rt | TAACATAAGGGACTGACCACCCGGG GCTTCA- TCATYAYACCAATCTGC |
| p121-F | CGTCTTCAAAGCAAGTGGATT |
| p121-R | CCAACGCTGATCAATTCCAC |
Table 1 Primer information of G. pentaphyllu GpmiR156a and GpmiR166b
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| GpmiR156a-F | TCTTYACWCAACWTCTCACCCA |
| GpmiR156a-R | GAGARAYCWGCATAWCTCAATMACC |
| GpmiR166b-F | CAGATACATGTACGAGGWGTTCA |
| GpmiR166b-R | GCTTCATCATYAYACCAATCTGC |
| GpmiR156a-Ft | TGGAGAGAACACGGGGGACTCTAGA TCTTYACWCAACWTCTCACCCA |
| GpmiR156a-Rt | TAACATAAGGGACTGACCACCCGGG GAGARAYCWGCATAWCTCAATMACC |
| GpmiR166b-Ft | TGGAGAGAACACGGGGGACTCTAGA CAGATA- CATGTACGAGGWGTTCA |
| GpmiR166b-Rt | TAACATAAGGGACTGACCACCCGGG GCTTCA- TCATYAYACCAATCTGC |
| p121-F | CGTCTTCAAAGCAAGTGGATT |
| p121-R | CCAACGCTGATCAATTCCAC |

Fig. 2 Precursor sequences of G. pentaphyllu GpmiR156a and GpmiR166b A:GpmiR156a precursor sequence. B:GpmiR166b precursor sequence. Underlines indicate mature sequences of GpmiR156a and GpmiR166b,respectively

Fig. 3 Secondary structures of G. pentaphyllu GpmiR156a and GpmiR166b A:Secondary structure of GpmiR156a. B:Secondary structure of GpmiR166b. The color change in the figure refers to the base pairing probability and the red ones refer to the highest probability
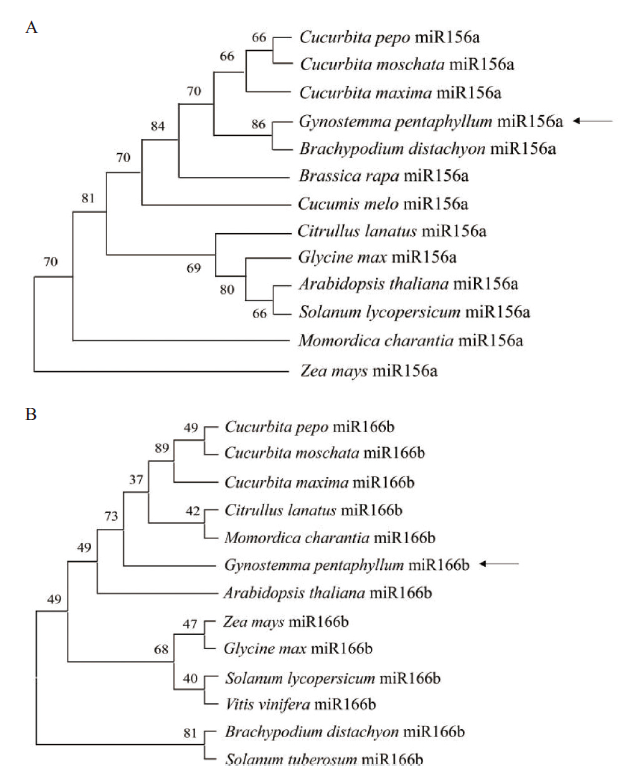
Fig. 4 Phylogenetic analysis of G. pentaphyllu GpmiR156a and GpmiR166b A:Phylogenetic analysis of GpmiR156a. B:Phylogenetic analysis of GpmiR166b. Arrows indicate the position of GpmiR156a and GpmiR166b,respectively
| GpmiR156a靶基因预测GpmiR156a target gene prediction | GpmiR166b靶基因预测GpmiR166b target gene prediction | |||
|---|---|---|---|---|
| 编号Serial No. | 名称Name | 编号Serial No. | 名称Name | |
| AT5G43270.3 | SPL2 | squamosa promoter binding protein-like 2 | AT1G30490.1 | PHV,ATHB9 | |
| AT2G33810.1 | SPL3 | squamosa promoter binding protein-like 3 | |||
| AT1G53160.1 | SPL4 | squamosa promoter binding protein-like 4 | AT1G52150.3 | ATHB-15,ATHB15,CNA | |
| AT3G57920.1 | SPL5 | squamosa promoter binding protein-like 5 | |||
| AT2G42200.1 | SPL9,AtSPL9 | squamosa promoter binding protein-like 9 | AT2G34710.1 | PHB,ATHB14,ATHB-14 | |
| AT1G27370.3 | SPL10 | squamosa promoter binding protein-like 10 | |||
| AT1G27360.4 | SPL11 | squamosa promoter-like 11 | AT4G32880.1 | ATHB-8,ATHB8,HB-8 | homeobox gene 8 | |
| AT5G50570.2 | SPL13A,SPL13 | Squamosa promoter-binding protein-like(SBP domain)transcription factor family protein | |||
| AT5G50670.1 | SPL13B,SPL13 | Squamosa promoter-binding protein-like(SBP domain)transcription factor family protein | AT5G60690.1 | REV,IFL | |
| AT3G57920.1 | SPL15 | squamosa promoter binding protein-like 15 | |||
Table 2 Prediction of G. pentaphyllu GpmiR156a and GpmiR166b target genes
| GpmiR156a靶基因预测GpmiR156a target gene prediction | GpmiR166b靶基因预测GpmiR166b target gene prediction | |||
|---|---|---|---|---|
| 编号Serial No. | 名称Name | 编号Serial No. | 名称Name | |
| AT5G43270.3 | SPL2 | squamosa promoter binding protein-like 2 | AT1G30490.1 | PHV,ATHB9 | |
| AT2G33810.1 | SPL3 | squamosa promoter binding protein-like 3 | |||
| AT1G53160.1 | SPL4 | squamosa promoter binding protein-like 4 | AT1G52150.3 | ATHB-15,ATHB15,CNA | |
| AT3G57920.1 | SPL5 | squamosa promoter binding protein-like 5 | |||
| AT2G42200.1 | SPL9,AtSPL9 | squamosa promoter binding protein-like 9 | AT2G34710.1 | PHB,ATHB14,ATHB-14 | |
| AT1G27370.3 | SPL10 | squamosa promoter binding protein-like 10 | |||
| AT1G27360.4 | SPL11 | squamosa promoter-like 11 | AT4G32880.1 | ATHB-8,ATHB8,HB-8 | homeobox gene 8 | |
| AT5G50570.2 | SPL13A,SPL13 | Squamosa promoter-binding protein-like(SBP domain)transcription factor family protein | |||
| AT5G50670.1 | SPL13B,SPL13 | Squamosa promoter-binding protein-like(SBP domain)transcription factor family protein | AT5G60690.1 | REV,IFL | |
| AT3G57920.1 | SPL15 | squamosa promoter binding protein-like 15 | |||

Fig. 5 Detection and phenotype analysis of positive transg-enic plants A:Partial PCR identification results of GpmiR156a and GpmiR166b transgenic lines. M:Marker. 1:Negative control. 2:Positive control of GpmiR156a. 3-12:GpmiR156a. 13:Positive control of GpmiR166b. 14-24:GpmiR166b. B:Identification of GpmiR156a and GpmiR166b transgenic lines by GUS staining. 1:Wild-type(WT)A. thaliana. 2-7:GpmiR156a transgenic plants. 8-13:GpmiR166b transgenic plants. Bar=5 mm. C:Kan resistance screening. a:GpmiR156a screening medium. b:GpmiR166b screening medium. Red arrows indicate the transgenic A. thaliana selected for Kan resistance. D:Phenotype of WT and transgenic A. thaliana. a,d:WT Arabidopsis thaliana. b,e:GpmiR156a transgenic A. thaliana. c,f:GpmiR166b transgenic A. thaliana. Bar=1 cm
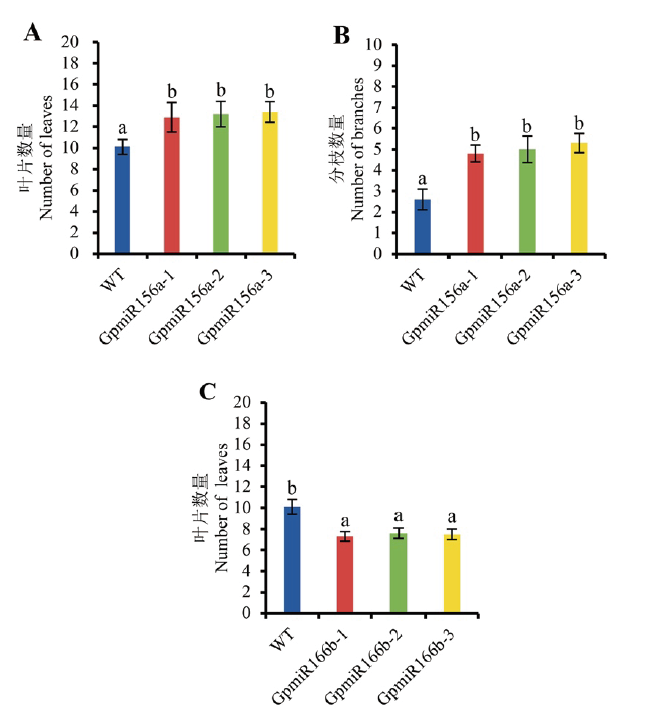
Fig. 6 Number of rosette leaves and branches in transgenic A. thaliana and WT A:Number of rosette leaves in the transgenic A. thaliana overexpressing GpmiR156a. B:Number of branches in the transgenic A. thaliana overexpressing GpmiR156a. C:Number of rosette leaves in the transgenic A. thaliana overexpressing GpmiR166b. GpmiR156a-1,GpmiR156a-2,and GpmiR156a-3 are three separate lines of transgenic A. thaliana. GpmiR166b-1,GpmiR166b-2,and GpmiR166b-3 are three separate lines of transgenic A. thaliana. Number of branches refers to the branches other than the main branches. n=10. Lowercase letters a and b refer to the significant difference between wild type and transgenic plants. The same below
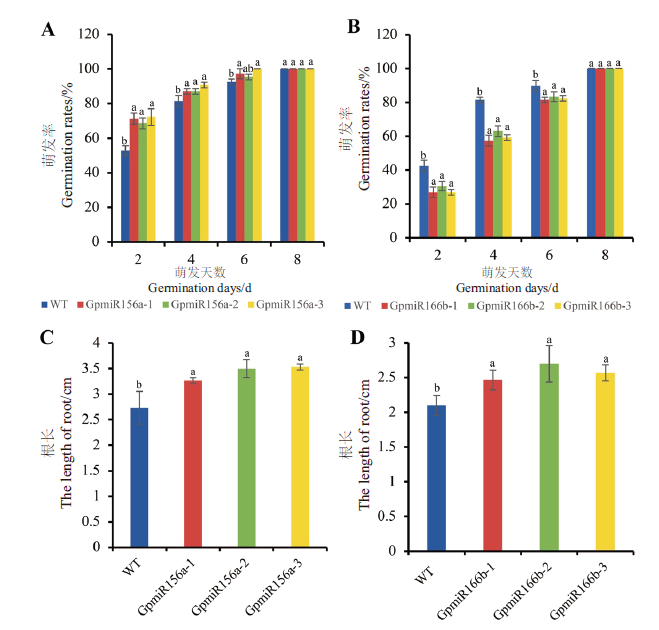
Fig. 7 Seed germination and root length of overexpressing GpmiR156a and GpmiR166b and wild-type A. thal-iana A:Seed germination of transgenic Arabidopsis overexpressing GpmiR156a. B:Seed germination of transgenic Arabidopsis overexpressing GpmiR166b. C:Root length of transgenic Arabidopsisoverexpressing GpmiR156a. D:Root length of transgenic Arabidopsis overexpressing GpmiR166b. GpmiR156a-1,GpmiR156a-2,and GpmiR156a-3 are three separate lines of transgenic Arabidopsis. GpmiR166b-1,GpmiR166b-2,and GpmiR166b-3 are three separate lines of transgenic Arabidopsis. A,B:n=36;C,D:n=5
| [1] |
Razmovski-Naumovski V, Huang THW, Tran VH, et al. Chemistry and pharmacology of Gynostemma pentaphyllum[J]. Phytochem Rev, 2005, 4(2/3):197-219.
doi: 10.1007/s11101-005-3754-4 URL |
| [2] |
Yang Q, Liu SB, Han XN, et al. Integrated transcriptome and miRNA analysis uncovers molecular regulators of aerial stem-to-rhizome transition in the medical herb Gynostemma pentaphyllum[J]. BMC Genomics, 2019, 20(1):865.
doi: 10.1186/s12864-019-6250-8 pmid: 31730459 |
| [3] |
Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets[J]. Trends Plant Sci, 2008, 13(7):343-349.
doi: 10.1016/j.tplants.2008.03.009 URL |
| [4] |
Ma JY, Zhao P, Liu SB, et al. The control of developmental phase transitions by microRNAs and their targets in seed plants[J]. Int J Mol Sci, 2020, 21(6):1971.
doi: 10.3390/ijms21061971 URL |
| [5] |
Schwab R, Palatnik JF, Riester M, et al. Specific effects of microRNAs on the plant transcriptome[J]. Dev Cell, 2005, 8(4):517-527.
doi: 10.1016/j.devcel.2005.01.018 URL |
| [6] |
Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity[J]. Nat Genet, 2007, 39(8):1033-1037.
pmid: 17643101 |
| [7] | He J, Xu ML, Willmann MR, et al. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana[J]. PLoS Genet, 2018, 14(4):e1007337. |
| [8] |
Wang J, Gao XY, Li L, et al. Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice[J]. J Exp Bot, 2010, 61(6):1885-1895.
doi: 10.1093/jxb/erp378 URL |
| [9] |
Williams L, Grigg SP, Xie MT, et al. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes[J]. Development, 2005, 132(16):3657-3668.
pmid: 16033795 |
| [10] |
Jung JH, Park CM. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis[J]. Planta, 2007, 225(6):1327-1338.
doi: 10.1007/s00425-006-0439-1 URL |
| [11] | Wang JW, Park MY, Wang LJ, et al. miRNA control of vegetative phase change in trees[J]. PLoS Genet, 2011, 7(2):e1002012. |
| [12] | Xiong JS, Bai YB, Ma CJ, et al. Molecular cloning and characterization of SQUAMOSA-promoter binding protein-like gene FvSPL10 from woodland strawberry(Fragaria vesca)[J]. Plants(Basel), 2019, 8(9):342. |
| [13] |
Guo Q, Li L, Zhao K, et al. Genome-wide analysis of poplar SQUAMOSA-promoter-binding protein(SBP)family under salt stress[J]. Forests, 2021, 12(4):413.
doi: 10.3390/f12040413 URL |
| [14] | Xu ML, Hu TQ, Zhao JF, et al. Developmental functions of mir156-regulated Squamosa promoter binding protein-like(spl)genes in Arabidopsis thaliana[J]. PLoS Genet, 2016, 12(8):e1006263. |
| [15] |
Gao RM, Gruber MY, Amyot L, et al. SPL13 regulates shoot branching and flowering time in Medicago sativa[J]. Plant Mol Biol, 2018, 96(1/2):119-133.
doi: 10.1007/s11103-017-0683-8 URL |
| [16] |
Miao CB, Wang Z, Zhang L, et al. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice[J]. Nat Commun, 2019, 10(1):3822.
doi: 10.1038/s41467-019-11830-5 URL |
| [17] |
Zhang L, Ding H, Jiang HL, et al. Regulation of cadmium tolerance and accumulation by miR156 in Arabidopsis[J]. Chemosphere, 2020, 242:125168.
doi: 10.1016/j.chemosphere.2019.125168 URL |
| [18] |
Barrera-Rojas CH, Rocha GHB, Polverari L, et al. miR156-targeted SPL10 controls Arabidopsis root meristem activity and root-derived de novo shoot regeneration via cytokinin responses[J]. J Exp Bot, 2020, 71(3):934-950.
doi: 10.1093/jxb/erz475 pmid: 31642910 |
| [19] |
Willmann MR, Poethig RS. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis[J]. Development, 2011, 138(4):677-685.
doi: 10.1242/dev.057448 pmid: 21228003 |
| [20] | Tang XR, Bian SM, Tang MJ, et al. microRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis[J]. PLoS Genet, 2012, 8(11):e1003091. |
| [21] |
Li ZX, Zhang LF, Li WF, et al. MIR166a affects the germination of somatic embryos in larixleptolepis by modulating IAA biosynthesis and signaling genes[J]. J Plant Growth Regul, 2017, 36(4):889-896.
doi: 10.1007/s00344-017-9693-7 URL |
| [22] |
Barik S, SarkarDas S, Singh A, et al. Phylogenetic analysis reveals conservation and diversification of micro RNA166 genes among diverse plant species[J]. Genomics, 2014, 103(1):114-121.
doi: 10.1016/j.ygeno.2013.11.004 URL |
| [23] |
Chaves I, Lin YC, Pinto-Ricardo C, et al. miRNA profiling in leaf and cork tissues of Quercus suber reveals novel miRNAs and tissue-specific expression patterns[J]. Tree Genet Genomes, 2014, 10(3):721-737.
doi: 10.1007/s11295-014-0717-1 URL |
| [24] |
Li ZX, Qi LW. Over-expression of LaMIR166a promotes organs development in Nicotiana benthamiana[J]. Russ J Plant Physiol, 2019, 66(5):718-724.
doi: 10.1134/S1021443719050133 URL |
| [25] | Gautam V, Singh A, Yadav S, et al. Conserved LBL1-ta-siRNA and miR165/166-RLD1/2 modules regulate root development in maize[J]. Development, 2021, 148(1):dev190033. |
| [1] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [2] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [3] | ZHANG Yu-juan, LI Dong-hua, GONG Hui-hui, CUI Xin-xiao, GAO Chun-hua, ZHANG Xiu-rong, YOU Jun, ZHAO Jun-sheng. Cloning and Salt-tolerance Analysis of NAC Transcription Factor SiNAC77 from Sesamum indicum L. [J]. Biotechnology Bulletin, 2023, 39(11): 308-317. |
| [4] | CHEN Hao-ting, ZHANG Yu-jing, LIU Jie, DAI Ze-min, LIU Wei, SHI Yu, ZHANG Yi, LI Tian-lai. Functional Analysis of WRKY6 Gene in Tomato Under Low-phosphorus Stress [J]. Biotechnology Bulletin, 2023, 39(10): 136-147. |
| [5] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [6] | CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(7): 51-61. |
| [7] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [8] | WANG Nan, ZHANG Rui, PAN Yang-yang, HE Hong-hong, WANG Jing-lei, CUI Yan, YU Si-jiu. Cloning of Bos grunniens TGF-β1 Gene and Its Expression in Major Organs of Female Reproductive System [J]. Biotechnology Bulletin, 2022, 38(6): 279-290. |
| [9] | LI Yu-hang, WANG Xing-ping, YANG Jian, LUORENG Zhuo-ma, REN Qian-qian, WEI Da-wei, MA Yun. Expression and Functional Analysis of miR-665 in Bovine Mammary Epithelial Cell Inflammation [J]. Biotechnology Bulletin, 2022, 38(5): 159-168. |
| [10] | LI Yang, ZHANG Xiao-tian, PIAO Jing-zi, ZHOU Ru-jun, LI Zi-bo, GUAN Hai-wen. Cloning and Bioinformatics Analysis of Blue-light Receptor EaWC 1 Gene in Elsinoë arachidis [J]. Biotechnology Bulletin, 2022, 38(5): 93-99. |
| [11] | ZHANG Lin, WEI Zhen-zhen, SONG Cheng-wei, GUO Li-li, GUO Qi, HOU Xiao-gai, WANG Hua-fang. Cloning and Expression Analysis of PoFD Gene from Paeonia ostii ‘Fengdan’ [J]. Biotechnology Bulletin, 2022, 38(11): 104-111. |
| [12] | ZHENG Qing-bo, YE Na, ZHANG Xiao-lan, BAO Peng-jia, WANG Fu-bin, REN Wen-wen, LIAO Yue-jiao, YAN Ping, PAN He-ping. Identification of Hair Follicle Cell Subsets and Bioinformatics Analysis of Characteristic Genes in Tianzhu White Yak During Catagen [J]. Biotechnology Bulletin, 2022, 38(10): 262-272. |
| [13] | FAN Ya-peng, RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei. Cloning,Expression and Preliminary Bioinformatics Analysis of the Alkaline Tolerant Gene GhZAT12 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 121-130. |
| [14] | DU Zhen-wei, ZHU Shuai-peng, MA Xiang-fei, LI Dong-hua, SUN Gui-rong. Cloning,Expression and Bioinformatics Analysis of the CDS Region of Chicken CEBPA Gene [J]. Biotechnology Bulletin, 2021, 37(8): 203-212. |
| [15] | HAO Xiang-yang, LIU Fan, WU Huan, WANG Bin, SUN Xue-li, XIANG Lei-lei, WANG Tian-chi, LAI Zhong-xiong, CHENG Chun-zhen. Cloning and Expression Analysis of GjPAL Genes in Gerbera jamesonni [J]. Biotechnology Bulletin, 2021, 37(6): 13-23. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||