








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (4): 278-286.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0813
Previous Articles Next Articles
YIN Liang1( ), WANG Dai-wei1,2, LIU Yue-ying1,2, LIU Hai-yan1, LUO Guang-hong1(
), WANG Dai-wei1,2, LIU Yue-ying1,2, LIU Hai-yan1, LUO Guang-hong1( )
)
Received:2023-08-20
Online:2024-04-26
Published:2024-04-30
Contact:
LUO Guang-hong
E-mail:yinl03@163.com;kyluo@hxu.edu.cn
YIN Liang, WANG Dai-wei, LIU Yue-ying, LIU Hai-yan, LUO Guang-hong. Cloning and Expression of Protease SpP1 Gene and Characterization of Enzymatic Properties[J]. Biotechnology Bulletin, 2024, 40(4): 278-286.
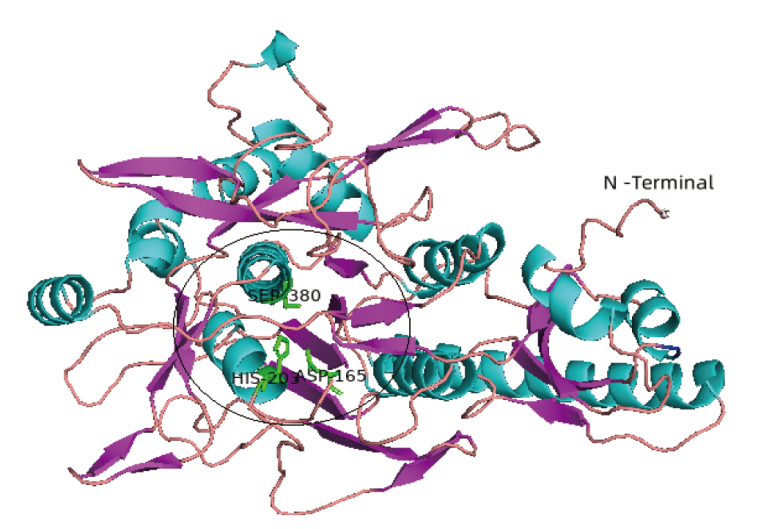
Fig. 1 3-D structure of peptidase SpP1 The 3D structure of SpP1 predicted by Swiss-Model. Pink indicates β-sheet; cyan indicates α-helix; orange indicates coil. The black circle shows the catalytic residues D165H203S380 and the substrate binding pocket
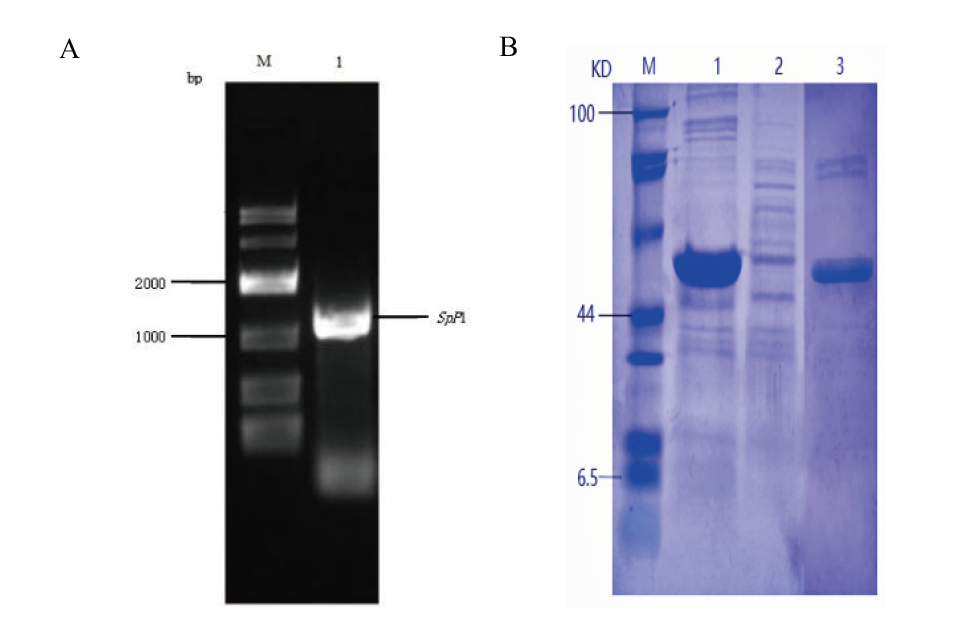
Fig. 2 SpP1 gel electrophoresis(A)and SDS - PAGE of SpP1(B) A: Gel electrophoresis map of SpP1 gene(M: DL10000 DNA marker. 1: PCR-amplified band of SpP1 gene). B: SDS-PAGE of SpP1protein(M: protein marker. 1: Precipitation. 2: Supernatant. 3: Purified SpP1 protein band)
| [1] | Ellaiah P, Srinivasulu B, Adinarayana K. A review on microbial alkaline proteases[J]. J Sci Ind Res, 2002, 61: 690-704. |
| [2] |
Gupta R, Beg QK, Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications[J]. Appl Microbiol Biotechnol, 2002, 59(1): 15-32.
doi: 10.1007/s00253-002-0975-y pmid: 12073127 |
| [3] | Matkawala F, Nighojkar S, Kumar A, et al. Microbial alkaline serine proteases: production, properties and applications[J]. World J Microbiol Biotechnol, 2021, 37(4): 63. |
| [4] |
Gurumallesh P, Alagu K, Ramakrishnan B, et al. A systematic reconsideration on proteases[J]. Int J Biol Macromol, 2019, 128: 254-267.
doi: S0141-8130(18)35199-7 pmid: 30664968 |
| [5] |
Contesini FJ, Melo RR, Sato HH. An overview of Bacillus proteases: from production to application[J]. Crit Rev Biotechnol, 2018, 38(3): 321-334.
doi: 10.1080/07388551.2017.1354354 pmid: 28789570 |
| [6] |
Yu P, Huang XX, Ren Q, et al. Purification and characterization of a H2O2-tolerant alkaline protease from Bacillus sp. ZJ1502, a newly isolated strain from fermented bean curd[J]. Food Chem, 2019, 274: 510-517.
doi: 10.1016/j.foodchem.2018.09.013 URL |
| [7] |
Dorra G, Ines K, Imen BS, et al. Purification and characterization of a novel high molecular weight alkaline protease produced by an endophytic Bacillus halotolerans strain CT2[J]. Int J Biol Macromol, 2018, 111: 342-351.
doi: 10.1016/j.ijbiomac.2018.01.024 URL |
| [8] |
Omrane Benmrad M, Moujehed E, Ben Elhoul M, et al. Production, purification, and biochemical characterization of serine alkaline protease from Penicillium chrysogenium strain X5 used as excellent bio-additive for textile processing[J]. Int J Biol Macromol, 2018, 119: 1002-1016.
doi: S0141-8130(18)33429-9 pmid: 30081129 |
| [9] |
Pérez-Lloréns JL, Benítez E, Vergara JJ, et al. Characterization of proteolytic enzyme activities in macroalgae[J]. Eur J Phycol, 2003, 38(1): 55-64.
doi: 10.1080/0967026031000096254 URL |
| [10] |
Lockau W, Massalsky B, Dirmeier A. Purification and partial characterization of a calcium-stimulated protease from the cyanobacterium, Anabaena variabilis[J]. Eur J Biochem, 1988, 172(2): 433-438.
pmid: 3127208 |
| [11] |
Strohmeier U, Gerdes C, Lockau W. Proteolysis in heterocyst-forming cyanobacteria: characterization of a further enzyme with trypsin-like specificity, and of a prolyl endopeptidase from Anabaena variabilis[J]. Z Naturforsch C J Biosci, 1994, 49(1-2): 70-78.
doi: 10.1515/znc-1994-1-212 pmid: 8148011 |
| [12] |
Niven GW. The characterization of two aminopeptidase activities from the cyanobacterium Anabaena flos-aquae[J]. Biochim Biophys Acta, 1995, 1253(2): 193-198.
pmid: 8519802 |
| [13] |
Nanni B, Balestreri E, Dainese E, et al. Characterisation of a specific phycocyanin-hydrolysing protease purified from Spirulina platensis[J]. Microbiol Res, 2001, 156(3): 259-266.
pmid: 11716214 |
| [14] |
Yada E, Nagata H, Noguchi Y, et al. An arginine specific protease from Spirulina platensis[J]. Mar Biotechnol, 2005, 7(5): 474-480.
doi: 10.1007/s10126-004-4115-9 URL |
| [15] | 吕冰心, 常蓉, 李博生. 基于蛋白质组学对螺旋藻在高温胁迫下响应机制的初步研究[J]. 植物生理学报, 2018, 54(5): 904-916. |
|
Lyu BX, Chang R, Li BS. Study on the response mechanism of Spirulina platensis to high temperature stress based on proteomics[J]. Plant Physiol J, 2018, 54(5): 904-916.
doi: 10.1104/pp.54.6.904 URL |
|
| [16] |
王娜, 向清豪, 赵肖荣, 等. 螺旋藻全蛋白与家族分类[J]. 食品科学, 2018, 39(16): 201-207.
doi: 10.7506/spkx1002-6630-201816029 |
| Wang N, Xiang QH, Zhao XR, et al. Family classification of the whole proteins of Spirulina[J]. Food Sci, 2018, 39(16): 201-207. | |
| [17] |
Gunes S, Tamburaci S, Dalay MC, et al. In vitro evaluation of Spirulina platensis extract incorporated skin cream with its wound healing and antioxidant activities[J]. Pharm Biol, 2017, 55(1): 1824-1832.
doi: 10.1080/13880209.2017.1331249 pmid: 28552036 |
| [18] | Stanier RY, Kunizawa MM, Cohen-Bazire G. Purification and property of unicellular blue-green algae (Order Chroococales)[J]. Bacteriol Rev, 1971, 35(2):120-171. |
| [19] |
Singh Chauhan R, Mani Mishra R. Characterization of alkaline protease producing Bacillus halodurans RSCVS-PF21 isolated from poultry farm soil[J]. Biosci, Biotech Res Asia, 2020, 17(2): 385-392.
doi: 10.13005/bbra/ URL |
| [20] |
Wen Y, Qiang J, Zhou G, et al. Characterization of redox and salinity-tolerant alkaline protease from Bacillus halotolerans strain DS5[J]. Front Microbiol, 2022, 13:935072.
doi: 10.3389/fmicb.2022.935072 URL |
| [21] | 李怡欣, 付刚, 马媛媛, 等. 碱性蛋白酶SubC在枯草芽孢杆菌中的高效异源表达[J]. 微生物学通报, 2021, 48(10): 3409-3420. |
| Li YX, Fu G, Ma YY, et al. Efficient heterologous expression of alkaline protease SubC in Bacillus subtilis[J]. Microbiol China, 2021, 48(10): 3409-3420. | |
| [22] | 周魏, 曾嵩玉, 余金凤, 等. 一株地衣芽胞杆菌产碱性蛋白酶条件优化[J]. 微生物学通报, 2022, 49(7): 2753-2766. |
| Zhou W, Zeng SY, Yu JF, et al. Optimization of alkaline protease production by a strain of Bacillus licheniformis[J]. Microbiol China, 2022, 49(7): 2753-2766. | |
| [23] | Hammami A, Bayoudh A, Hadrich B, et al. Response-surface methodology for the production and the purification of a new H2 O2-tolerant alkaline protease from Bacillus invictae AH1 strain[J]. Biotechnol Prog, 2020, 36(3): e2965. |
| [24] |
Meena P, Tripathi AD, Srivastava SK, et al. Utilization of agro-industrial waste(wheat bran)for alkaline protease production by Pseudomonas aeruginosa in SSF using Taguchi(DOE)methodology[J]. Biocatal Agric Biotechnol, 2013, 2(3): 210-216.
doi: 10.1016/j.bcab.2013.05.003 URL |
| [25] |
Boulkour Touioui S, Zaraî Jaouadi N, Bouacem K, et al. Biochemical and molecular characterization of a novel metalloprotease from Pseudomonas fluorescens strain TBS09[J]. Int J Biol Macromol, 2018, 107(Pt B): 2351-2363.
doi: S0141-8130(17)33230-0 pmid: 29055705 |
| [26] | Jenitta XJ, Priya S, Gnanadoss JJ. Optimization of culture conditions and inducers for improved protease production by Penicillium griseofulvum LCJ231 under submerged fermentation[J]. Int J Adv Biotechnol Res, 2015, 6(2):152-160. |
| [27] |
de Souza PM, Bittencourt ML, Caprara CC, et al. A biotechnology perspective of fungal proteases[J]. Braz J Microbiol, 2015, 46(2): 337-346.
doi: 10.1590/S1517-838246220140359 pmid: 26273247 |
| [28] |
Sattar H, Bibi Z, Kamran A, et al. Degradation of complex casein polymer: production and optimization of a novel serine metalloprotease from Aspergillus niger KIBGE-IB36[J]. Biocatal Agric Biotechnol, 2019, 21: 101256.
doi: 10.1016/j.bcab.2019.101256 URL |
| [29] |
Azrin NAM, Ali MSM, Rahman RNZRA, et al. Versatility of subtilisin: a review on structure, characteristics, and applications[J]. Biotechnol Appl Biochem, 2022, 69(6): 2599-2616.
doi: 10.1002/bab.v69.6 URL |
| [30] | 万明铼. 高产碱性蛋白酶菌株的筛选及酶学性质的研究[D]. 兰州: 兰州理工大学, 2023. |
| Wan ML. Screening of the bacterial strain producing high level of alkaline protease and study on its enzymatic properties[D]. Lanzhou: Lanzhou University of Technology, 2023. | |
| [31] |
Grøn H, Bech LM, Sørensen SB, et al. Studies of binding sites in the subtilisin from Bacillus lentus by means of site directed mutagenesis and kinetic investigations[J]. Adv Exp Med Biol, 1996, 379: 105-112.
pmid: 8796314 |
| [32] |
Kumar CG, Joo HS, Koo YM, et al. Thermostable alkaline protease from a novel marine haloalkalophilic Bacillus clausii isolate[J]. World J Microbiol Biotechnol, 2004, 20(4): 351-357.
doi: 10.1023/B:WIBI.0000033057.28828.a7 URL |
| [33] | Akel H, Yousef T. Characterization of a purified thermostable protease from hyperthermophilic Bacillus strain HUTBS71[J]. Eur J Sci Res, 2009, 31(2):280-288. |
| [34] |
Mei CF, Jiang XL. A novel surfactant- and oxidation-stable alkaline protease from Vibrio metschnikovii DL 33-51[J]. Process Biochem, 2005, 40(6): 2167-2172.
doi: 10.1016/j.procbio.2004.08.007 URL |
| [35] |
Jang JS, Kang DO, Chun MJ, et al. Molecular cloning of a subtilisin J gene from Bacillus stearothermophilus and its expression in Bacillus subtilis[J]. Biochem Biophys Res Commun, 1992, 184(1): 277-282.
doi: 10.1016/0006-291X(92)91189-W URL |
| [36] |
Neidhart DJ, Petsko GA. The refined crystal structure of subtilisin Carlsberg at 2.5 A resolution[J]. Protein Eng, 1988, 2(4): 271-276.
pmid: 3150541 |
| [37] |
Nonaka T, Suzuki T, Tanaka N, et al. Structure and function of subtilisin BPN'as studied through crystallographic studies on a series of its complexes with genetically engineered proteinaceous inhibitor SSI[J]. Adv Exp Med Biol, 1996, 379: 21-27.
pmid: 8796307 |
| [38] |
Gaur S, Agrahari S, Wadhwa N. Purification of protease from Pseudomonas thermaerum GW1 isolated from poultry waste site[J]. Open Microbiol J, 2010, 4: 67-74.
doi: 10.2174/1874285801004010067 URL |
| [39] |
Jellouli K, Ghorbel-Bellaaj O, Ben Ayed H, et al. Alkaline-protease from Bacillus licheniformis MP1: purification, characterization and potential application as a detergent additive and for shrimp waste deproteinization[J]. Process Biochem, 2011, 46(6): 1248-1256.
doi: 10.1016/j.procbio.2011.02.012 URL |
| [40] |
Sousa F, Jus S, Erbel A, et al. A novel metalloprotease from Bacillus cereus for protein fibre processing[J]. Enzyme Microb Technol, 2007, 40(7): 1772-1781.
doi: 10.1016/j.enzmictec.2006.12.017 URL |
| [1] | ZHENG Fei, YANG Jun-zhao, NIU Yu-feng, LI Rui-lin, ZHAO Guo-zhu. Characterization and Functional Analysis of Lytic Polysaccharide Monooxygenase TtLPMO9I from Thermothelomyces thermophilus [J]. Biotechnology Bulletin, 2024, 40(2): 289-299. |
| [2] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [3] | MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose [J]. Biotechnology Bulletin, 2023, 39(4): 124-135. |
| [4] | CHEN Nan-nan, WANG Chun-lai, JIANG Zhen-zhong, JIAO Peng, GUAN Shu-yan, MA Yi-yong. Genetic Transformation and Chilling Resistance Analysis of Maize ZmDHN15 Gene in Tobacco [J]. Biotechnology Bulletin, 2023, 39(4): 259-267. |
| [5] | YANG Jun-zhao, ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei. Structure and Function Analysis of Novel GH5 Multi-domain Cellulase [J]. Biotechnology Bulletin, 2023, 39(4): 71-80. |
| [6] | XING Yuan, SONG Jian, LI Jun-yi, ZHENG Ting-ting, LIU Si-chen, QIAO Zhi-jun. Identification of AP Gene Family and Its Response Analysis to Abiotic Stress in Setaria italica [J]. Biotechnology Bulletin, 2023, 39(11): 238-251. |
| [7] | YIN Guo-ying, LIU Chang, CHANG Yong-chun, YU Wang-jie, WANG Bing, ZHANG Pan, GUO Yu-shuang. Identification of the Cysteine Protease Family and Corresponding miRNAs in Nicotiana tabacum L. and Their Responses to PVY [J]. Biotechnology Bulletin, 2023, 39(10): 184-196. |
| [8] | WANG Yu-chen, DING Zun-dan, GUAN Fei-fei, TIAN Jian, LIU Guo-an, WU Ning-feng. Identification of the Thermostable Laccase Gene ba4 and Characterization of Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(8): 252-260. |
| [9] | FU Qiao, LIN Qi-lan, XUE Qiang, XIONG Hai-rong, WANG Ya-wei. Effects of CBM41 N-terminal Truncation on the Enzymological Properties of the Pullulanase from Bacillus subtilis 168 [J]. Biotechnology Bulletin, 2022, 38(6): 245-251. |
| [10] | NIU Xin, ZHANG Ying, WANG Mao-jun, LIU Wen-long, LU Fu-ping, LI Yu. Effects of Different Integration Sites on the Expression of Exogenous Alkaline Protease in Bacillus amyloliquefaciens [J]. Biotechnology Bulletin, 2022, 38(4): 253-260. |
| [11] | WANG Bo-ya, JIANG Yong, HUANG Yan, CAO Ying, HU Shang-lian. Cloning and Functional Analysis of BeCesA4 in Bambusa emeiensis [J]. Biotechnology Bulletin, 2022, 38(11): 185-193. |
| [12] | WANG Xiao-tao, ZOU Hang, WU Yi, XIANG Shen-wei, LV Hua, LIU Chao-lan, LIN Jia-fu, WANG Xin-rong, CHU Yi-wen, SONG Tao. Heterologous Expression and Enzymatic Properties Analysis of Novel β-agarase Aga2 from Paraglaciecola hydrolytica [J]. Biotechnology Bulletin, 2022, 38(11): 258-268. |
| [13] | ZHANG Tong-tong, ZHENG Deng-yu, WU Zhong-yi, ZHANG Zhong-bao, YU Rong. Functional Analysis of ZmNF-YB13 Responding to Drought and Salt Stress [J]. Biotechnology Bulletin, 2022, 38(10): 115-123. |
| [14] | SU Yu, LI Zong-yun, HAN Yong-hua. Advances in Plant Vacuolar Processing Enzymes [J]. Biotechnology Bulletin, 2021, 37(6): 181-191. |
| [15] | MO Li-jie, LIU Xia-tong, LI Hui, LU Hai. On the Function of Plant Cysteine Protease in Plant Growth and Development [J]. Biotechnology Bulletin, 2021, 37(6): 202-212. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||