








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 5-22.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1186
Previous Articles Next Articles
HU Ya-dan1( ), WU Guo-qiang1(
), WU Guo-qiang1( ), LIU Chen2, WEI Ming1
), LIU Chen2, WEI Ming1
Received:2023-12-15
Online:2024-06-26
Published:2024-05-14
Contact:
WU Guo-qiang
E-mail:huyadan001003@163.com;gqwu@lut.edu.cn
HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress[J]. Biotechnology Bulletin, 2024, 40(6): 5-22.
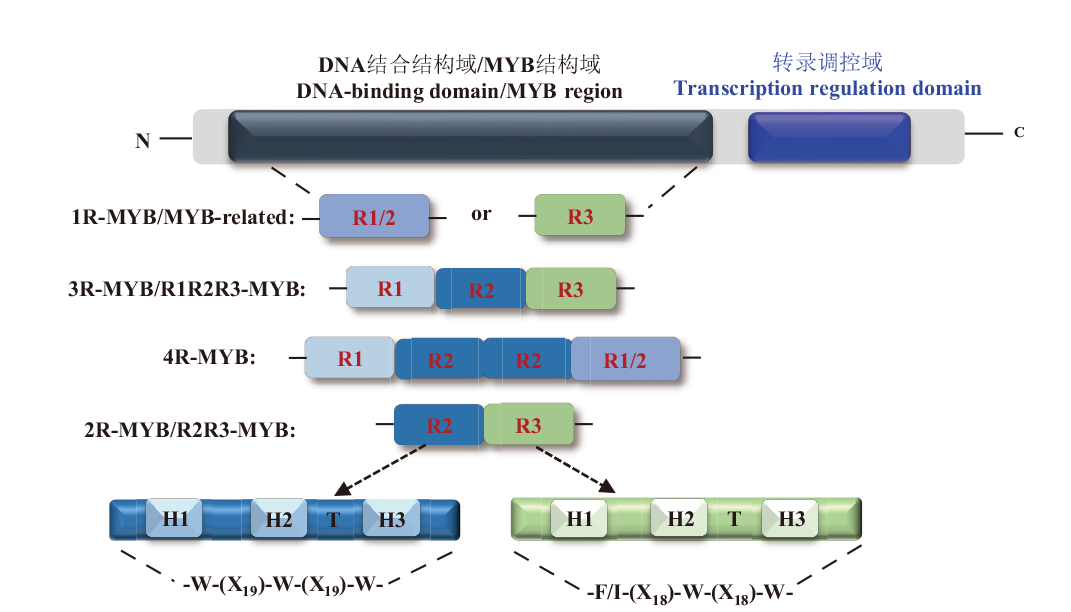
Fig. 1 Domain structure of MYB transcription factors R1, R2, and R3 are the MYB domain, where H1-H3 indicate the α-helix and T indicate the β-turn. W: Trp; F: Phe; I: Ile; X: amino acid
| 物种 Species | 基因名称Gene name | 基因总数Number of total genes | 分类Classification | 参考文献Reference | |||
|---|---|---|---|---|---|---|---|
| R2R3-MYB | R1R2R3-MYB | MYB-related | Atypical MYB | ||||
| 拟南芥Arabidopsis thaliana | AtMYB | 198 | 126 | 5 | 64 | 3 | [ |
| 水稻Oryza sativa | OsMYB | 190 | 99 | 3 | 87 | 1 | [ |
| 大豆Glycine max | GmMYB | 254 | 244 | 6 | / | 4 | [ |
| 甜菜Beta vulgaris | BvMYB | 75 | 70 | 3 | / | 2 | [ |
| 番茄Solanum lycopersicum | SlMYB | 127 | 122 | 4 | / | 1 | [ |
| 矮牵牛Petunia hybrida | PaMYB | 155 | 106 | 7 | 40 | 2 | [ |
| 猕猴桃Actinidia chinensis | AcMYB | 181 | 91 | 3 | 87 | / | [ |
| 辣椒Capsicum annuum | CaMYB | 215 | 116 | 5 | 92 | 2 | [ |
| 小果野蕉Musa acuminata | MaMYB | 305 | 222 | 7 | 73 | 3 | [ |
| 野蕉Musa balbisiana | MbMYB | 251 | 184 | 5 | 59 | 3 | [ |
| 火龙果Hylocereus undatus | HuMYB | 185 | 105 | 4 | 75 | 1 | [ |
| 马铃薯Solanum tuberosum | StMYB | 217 | 124 | 3 | 90 | / | [ |
| 萝卜Raphanus sativus | RsMYB | 187 | 174 | 9 | 2 | 2 | [ |
| 铁皮石斛Dendrobium officinale | DoMYB | 164 | 117 | 4 | 42 | 1 | [ |
| 花生Arachis hypogaea | AhMYB | 443 | 209 | 12 | 219 | 3 | [ |
| 龙眼Dimocarpus longan | DlMYB | 219 | 119 | 3 | 95 | 2 | [ |
| 甘蓝型油菜Brassica napus | BnMYB | 680 | 429 | 22 | 227 | 2 | [ |
| 木麻黄Casuarina equisetifolia | CeqMYB | 182 | 107 | 4 | 69 | 2 | [ |
| 甜樱桃Prunus avium | PavMYB | 69 | 14 | 2 | 51 | 2 | [ |
Table 1 MYB genes in different plant species
| 物种 Species | 基因名称Gene name | 基因总数Number of total genes | 分类Classification | 参考文献Reference | |||
|---|---|---|---|---|---|---|---|
| R2R3-MYB | R1R2R3-MYB | MYB-related | Atypical MYB | ||||
| 拟南芥Arabidopsis thaliana | AtMYB | 198 | 126 | 5 | 64 | 3 | [ |
| 水稻Oryza sativa | OsMYB | 190 | 99 | 3 | 87 | 1 | [ |
| 大豆Glycine max | GmMYB | 254 | 244 | 6 | / | 4 | [ |
| 甜菜Beta vulgaris | BvMYB | 75 | 70 | 3 | / | 2 | [ |
| 番茄Solanum lycopersicum | SlMYB | 127 | 122 | 4 | / | 1 | [ |
| 矮牵牛Petunia hybrida | PaMYB | 155 | 106 | 7 | 40 | 2 | [ |
| 猕猴桃Actinidia chinensis | AcMYB | 181 | 91 | 3 | 87 | / | [ |
| 辣椒Capsicum annuum | CaMYB | 215 | 116 | 5 | 92 | 2 | [ |
| 小果野蕉Musa acuminata | MaMYB | 305 | 222 | 7 | 73 | 3 | [ |
| 野蕉Musa balbisiana | MbMYB | 251 | 184 | 5 | 59 | 3 | [ |
| 火龙果Hylocereus undatus | HuMYB | 185 | 105 | 4 | 75 | 1 | [ |
| 马铃薯Solanum tuberosum | StMYB | 217 | 124 | 3 | 90 | / | [ |
| 萝卜Raphanus sativus | RsMYB | 187 | 174 | 9 | 2 | 2 | [ |
| 铁皮石斛Dendrobium officinale | DoMYB | 164 | 117 | 4 | 42 | 1 | [ |
| 花生Arachis hypogaea | AhMYB | 443 | 209 | 12 | 219 | 3 | [ |
| 龙眼Dimocarpus longan | DlMYB | 219 | 119 | 3 | 95 | 2 | [ |
| 甘蓝型油菜Brassica napus | BnMYB | 680 | 429 | 22 | 227 | 2 | [ |
| 木麻黄Casuarina equisetifolia | CeqMYB | 182 | 107 | 4 | 69 | 2 | [ |
| 甜樱桃Prunus avium | PavMYB | 69 | 14 | 2 | 51 | 2 | [ |

Fig. 2 Phylogenetic tree of MYB family in higher plants MEGA 11 software was used for sequence multiple comparisons and phylogenetic tree construction. Blue circles indicate A. thaliana; red stars indicate B. vulgaris. The source, name, and accession number of MYBs are shown in the supplementary Table 1
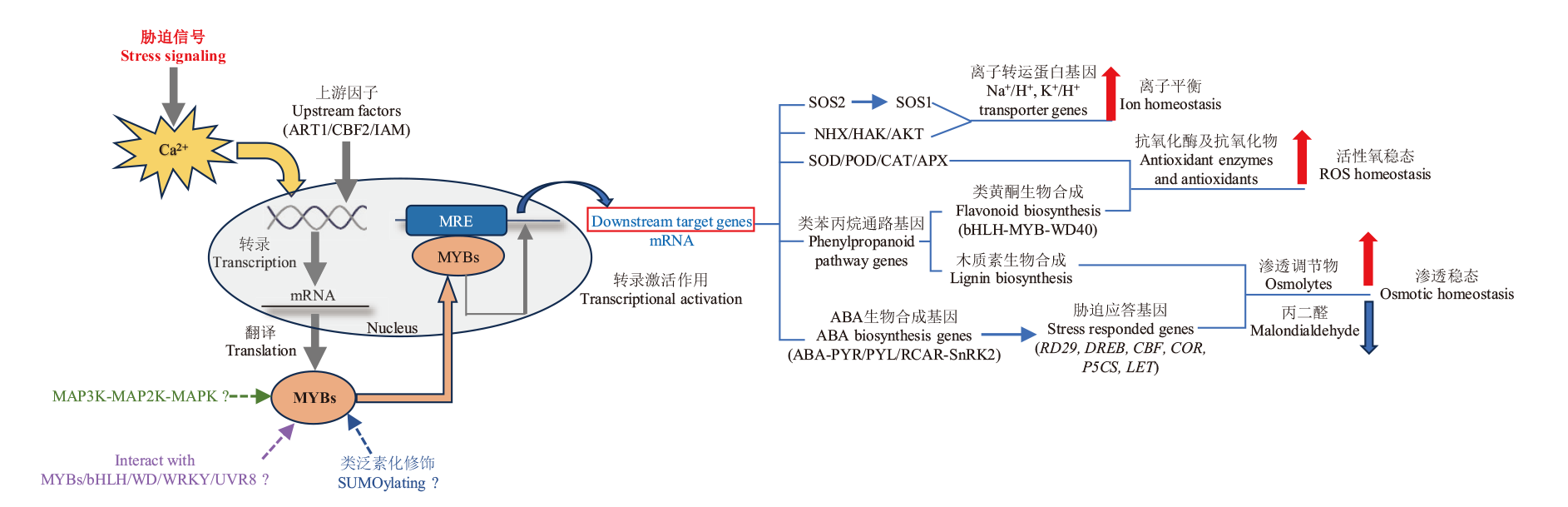
Fig. 3 MYB transcription factors involved in regulating the responses to various stresses in plants Solid arrows indicate direct interactions or activations, while dotted lines and question marks indicate that they occur only in certain MYB regulatory pathways and are not widespread. Bold red and blue arrows indicate up-regulation and down-regulation respectively
| 物种 Species | 基因名称 Gene name | 基因亚家族 Gene subfamily | 靶基因及位点 Target genes and sites | 功能 Function | 胁迫响应 Stress responses | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 拟南芥 Arabidopsis thaliana | AtMYB25 | R2R3-MYB | DREB2C, RD29a, SLAH1, JAZ10 | 激活下游胁迫应答基因 | 盐分、渗透性、脱落酸 | [ |
| AtMYB37 | R2R3-MYB | ABF2/3, COR15A, RD29a, RD22, PSII/I | 提高PSII活性,调节能量耗散比例 | 盐分、脱落酸、干旱 | [ | |
| AtMYBS1 | MYB-related | MAX1 | 负向调控独脚金素内酯途径 | 高温(-) | [ | |
| AtMYB74 | R2R3-MYB | ERF53, NIG1, HSFA6a, MYB47, MYB90, MYB102 | 受生长素前体IAM激活后调控下游靶基因 | 渗透、高温 | [ | |
| AtMYB12 | R2R3-MYB | Flavonoid biosynthesis genes, ZEP, NCED, ABA2, AAO, P5CS, P5CR, LEA, SOD, CAT, POD | 提高胁迫条件下植物中黄酮类化合物含量 | 盐分、干旱、高温、紫外线 | [ | |
| AtMYB71 | R2R3-MYB | ABA response genes | 调节植物ABA应答 | 脱落酸 | [ | |
| AtMYB94/96 | R2R3-MYB | KCS1/2/6, KCR1, CER1/3, WSD1 | 促进植物表皮蜡质生物合成 | 干旱、强光、紫外线 | [ | |
| 花生 Arachis hypogaea | AhMYB30 | MYB-related | KIN1, COR15a, RD29A, ABI2 | 上调参与DREB/CBF和ABA信号途径的下游胁迫相关基因表达 | 低温、盐分 | [ |
| 白菜型油菜 Brassica campestris | BcMYB111 | R2R3-MYB | F3H, FLS1 | 被CBF转录激活后增强黄酮类化合物生物合成 | 低温 | [ |
| 水稻 Oryza sativa | OsMYBR57 | MYB-related | OsbZIPs transcription is regulated after interaction with OsHB22 | 与HB22互作后激活转录因子bZIP | 干旱 | [ |
| OsMYB-R1 | MYB-related | SOD, CAT, GPX, LEA, ABRE | 激活下游胁迫相关基因 | 干旱、铬元素、水杨酸、脱落酸、茉莉酸 | [ | |
| OsFLP | R2R3-MYB | NAC1/6, DST, peroxidase 24 precursor | 激活下游相关转录因子 | 脱落酸、干旱、盐分 | [ | |
| 陆地棉 Gossypium hirsutum | GhMYB36 | R2R3-MYB | PR1 | 激活下游胁迫相关基因 | 干旱、黄萎病 | [ |
| GhMYB102 | R2R3-MYB | NCED1, ZAT10 | 参与调控ABA生物合成及干旱应答基因表达 | 干旱 | [ | |
| 二色补血草 Limonium bicolor | LbMYB48 | MYB-related | CPC-like, DIS3, SOSs, RLKs, GSTs | 调控表皮发育相关及盐胁迫相关基因表达 | 盐分 | [ |
| LbTRY | MYB-related | GL3, ZFP5, RHD6, RSL1, LRL2/3, SOS1/2/3, P5CS | 上调GL3/ZFP5表达后与表达产物竞争性结合,改变转基因植株表皮细胞分化方向,使得根毛的发育增强,吸收更多的Na+ | 盐分(-) | [ | |
| 山核桃 Carya cathayensis | CcMYB12 | R2R3-MYB | C4H, CHI, F3H, ANR, ANS, DFR | 参与花青素合成途径 | 盐分、干旱、酸 | [ |
| 橡胶树 Hevea brasiliensis | HbMYB44 | R2R3-MYB | Homologous genes and interacting protein-encoding genes | 激活下游相关基因 | 盐分、渗透、干旱、脱落酸、茉莉酸甲酯、赤霉素、水杨酸 | [ |
| 甘薯 Ipomoea batatas | IbMYB308 | R2R3-MYB | SOD, POD, APX, P5CS | 激活下游胁迫相关基因 | 盐分 | [ |
| IbMYB73 | R2R3-MYB | NCED3, ABA2, AAO3, ABI2, SnRK2.3, DREB1D, RD22, RD26, GER5 | 激活ABA依赖型的不定根生长及胁迫耐受性的负调控因子转录表达 | 脱落酸(-)、盐分(-)、干旱(-) | [ | |
| 灰绿藜 Chenopodium glaucum | CgMYB1 | R2R3-MYB | NHX1, HAK5, SOS1, P5CS2, SOD, POD1, CBF1, COR15, COR47, bHLH001 | 提高转基因植物生理机能及胁迫相关基因表达 | 盐分、低温 | [ |
| 豌豆 Pisum sativum | PsFLP | R2R3-MYB | CYCA2;3, CDKA;1, AAO3, NCED3, SnRK2.3 | 调控气孔形成及ABA合成与信号转导基因 | 脱落酸、干旱 | [ |
| 84K杨 Populus alba × Populus glandulosa | PagMYB205 | R2R3-MYB | POD, SOD, CAT and root vitality related genes | 负调节抗氧化酶活性及根活力 | 盐分(-) | [ |
| PagMYB151 | R2R3-MYB | Proline biosynthesis genes | 与共表达转录因子共同改变根结构,促进脯氨酸积累及减少MDA含量 | 盐分 | [ | |
| 葡萄 V. labrusca×V. riparia | VhMYB2 | R2R3-MYB | SOS1/2/3, NHX1, SnRK2.6, NCED3, P5CS1, CAT1 | 激活下游相关基因 | 高盐、干旱 | [ |
| 辣椒 Capsicum annuum | CaDIM1 | MYB-related | OSR1, RAB18, NCED3 and stress-responsive genes | 诱导胁迫/ABA相关基因表达 | 脱落酸、干旱 | [ |
| 荞麦 Fagopyrum tataricum | FtMYB11 | R2R3-MYB | CBF1, DREB2A, RD20, ABA3, NCED3, C4H, 4CL, F3H, ANS, DFR | 调节ABA信号途径、干旱及类黄酮生物合成相关基因表达水平 | 脱落酸、干旱(-)、盐分(-) | [ |
| 芹菜 Apium graveolens | AgMYB5 | R2R3-MYB | CRTISO, LCYB, ABA1/2, NCED6, AAO3, ERD1, RD22, P5CS1, RD29 | 增强了β-胡萝卜素生物合成,随后诱导ABA合成 | 氧化损伤、干旱 | [ |
| 梁 Setaria italica | SiMYB16 | MYB-related | CSE, FAR1, CYP87A3, 4CL1, PAL, F5H, COMT, NCED3 | 调控植物木质素、类黄酮及木栓质生物合成 | 盐分 | [ |
| 木薯 Manihot esculenta | MeMYB60 | R2R3-MYB | CAT1/2 | 参与保卫细胞中活性氧稳态并影响气孔运动 | 脱落酸、干旱 | [ |
| 葡萄 Vitis amurensis | VaMYB14 | R2R3-MYB | ABA signaling genes, CORs, LTPs, CAT, POD | 参与激活ABA信号组分和CBF-COR非依赖的LTP3表达 | 盐分、低温、干旱 | [ |
| 番茄 Solanum lycopersicum | SlMYB41 | R2R3-MYB | SlHSP90.3 | 维持热胁迫下的活性氧稳态 | 高温 | [ |
Table 2 Function of MYB in regulating the response of plants to abiotic stress
| 物种 Species | 基因名称 Gene name | 基因亚家族 Gene subfamily | 靶基因及位点 Target genes and sites | 功能 Function | 胁迫响应 Stress responses | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 拟南芥 Arabidopsis thaliana | AtMYB25 | R2R3-MYB | DREB2C, RD29a, SLAH1, JAZ10 | 激活下游胁迫应答基因 | 盐分、渗透性、脱落酸 | [ |
| AtMYB37 | R2R3-MYB | ABF2/3, COR15A, RD29a, RD22, PSII/I | 提高PSII活性,调节能量耗散比例 | 盐分、脱落酸、干旱 | [ | |
| AtMYBS1 | MYB-related | MAX1 | 负向调控独脚金素内酯途径 | 高温(-) | [ | |
| AtMYB74 | R2R3-MYB | ERF53, NIG1, HSFA6a, MYB47, MYB90, MYB102 | 受生长素前体IAM激活后调控下游靶基因 | 渗透、高温 | [ | |
| AtMYB12 | R2R3-MYB | Flavonoid biosynthesis genes, ZEP, NCED, ABA2, AAO, P5CS, P5CR, LEA, SOD, CAT, POD | 提高胁迫条件下植物中黄酮类化合物含量 | 盐分、干旱、高温、紫外线 | [ | |
| AtMYB71 | R2R3-MYB | ABA response genes | 调节植物ABA应答 | 脱落酸 | [ | |
| AtMYB94/96 | R2R3-MYB | KCS1/2/6, KCR1, CER1/3, WSD1 | 促进植物表皮蜡质生物合成 | 干旱、强光、紫外线 | [ | |
| 花生 Arachis hypogaea | AhMYB30 | MYB-related | KIN1, COR15a, RD29A, ABI2 | 上调参与DREB/CBF和ABA信号途径的下游胁迫相关基因表达 | 低温、盐分 | [ |
| 白菜型油菜 Brassica campestris | BcMYB111 | R2R3-MYB | F3H, FLS1 | 被CBF转录激活后增强黄酮类化合物生物合成 | 低温 | [ |
| 水稻 Oryza sativa | OsMYBR57 | MYB-related | OsbZIPs transcription is regulated after interaction with OsHB22 | 与HB22互作后激活转录因子bZIP | 干旱 | [ |
| OsMYB-R1 | MYB-related | SOD, CAT, GPX, LEA, ABRE | 激活下游胁迫相关基因 | 干旱、铬元素、水杨酸、脱落酸、茉莉酸 | [ | |
| OsFLP | R2R3-MYB | NAC1/6, DST, peroxidase 24 precursor | 激活下游相关转录因子 | 脱落酸、干旱、盐分 | [ | |
| 陆地棉 Gossypium hirsutum | GhMYB36 | R2R3-MYB | PR1 | 激活下游胁迫相关基因 | 干旱、黄萎病 | [ |
| GhMYB102 | R2R3-MYB | NCED1, ZAT10 | 参与调控ABA生物合成及干旱应答基因表达 | 干旱 | [ | |
| 二色补血草 Limonium bicolor | LbMYB48 | MYB-related | CPC-like, DIS3, SOSs, RLKs, GSTs | 调控表皮发育相关及盐胁迫相关基因表达 | 盐分 | [ |
| LbTRY | MYB-related | GL3, ZFP5, RHD6, RSL1, LRL2/3, SOS1/2/3, P5CS | 上调GL3/ZFP5表达后与表达产物竞争性结合,改变转基因植株表皮细胞分化方向,使得根毛的发育增强,吸收更多的Na+ | 盐分(-) | [ | |
| 山核桃 Carya cathayensis | CcMYB12 | R2R3-MYB | C4H, CHI, F3H, ANR, ANS, DFR | 参与花青素合成途径 | 盐分、干旱、酸 | [ |
| 橡胶树 Hevea brasiliensis | HbMYB44 | R2R3-MYB | Homologous genes and interacting protein-encoding genes | 激活下游相关基因 | 盐分、渗透、干旱、脱落酸、茉莉酸甲酯、赤霉素、水杨酸 | [ |
| 甘薯 Ipomoea batatas | IbMYB308 | R2R3-MYB | SOD, POD, APX, P5CS | 激活下游胁迫相关基因 | 盐分 | [ |
| IbMYB73 | R2R3-MYB | NCED3, ABA2, AAO3, ABI2, SnRK2.3, DREB1D, RD22, RD26, GER5 | 激活ABA依赖型的不定根生长及胁迫耐受性的负调控因子转录表达 | 脱落酸(-)、盐分(-)、干旱(-) | [ | |
| 灰绿藜 Chenopodium glaucum | CgMYB1 | R2R3-MYB | NHX1, HAK5, SOS1, P5CS2, SOD, POD1, CBF1, COR15, COR47, bHLH001 | 提高转基因植物生理机能及胁迫相关基因表达 | 盐分、低温 | [ |
| 豌豆 Pisum sativum | PsFLP | R2R3-MYB | CYCA2;3, CDKA;1, AAO3, NCED3, SnRK2.3 | 调控气孔形成及ABA合成与信号转导基因 | 脱落酸、干旱 | [ |
| 84K杨 Populus alba × Populus glandulosa | PagMYB205 | R2R3-MYB | POD, SOD, CAT and root vitality related genes | 负调节抗氧化酶活性及根活力 | 盐分(-) | [ |
| PagMYB151 | R2R3-MYB | Proline biosynthesis genes | 与共表达转录因子共同改变根结构,促进脯氨酸积累及减少MDA含量 | 盐分 | [ | |
| 葡萄 V. labrusca×V. riparia | VhMYB2 | R2R3-MYB | SOS1/2/3, NHX1, SnRK2.6, NCED3, P5CS1, CAT1 | 激活下游相关基因 | 高盐、干旱 | [ |
| 辣椒 Capsicum annuum | CaDIM1 | MYB-related | OSR1, RAB18, NCED3 and stress-responsive genes | 诱导胁迫/ABA相关基因表达 | 脱落酸、干旱 | [ |
| 荞麦 Fagopyrum tataricum | FtMYB11 | R2R3-MYB | CBF1, DREB2A, RD20, ABA3, NCED3, C4H, 4CL, F3H, ANS, DFR | 调节ABA信号途径、干旱及类黄酮生物合成相关基因表达水平 | 脱落酸、干旱(-)、盐分(-) | [ |
| 芹菜 Apium graveolens | AgMYB5 | R2R3-MYB | CRTISO, LCYB, ABA1/2, NCED6, AAO3, ERD1, RD22, P5CS1, RD29 | 增强了β-胡萝卜素生物合成,随后诱导ABA合成 | 氧化损伤、干旱 | [ |
| 梁 Setaria italica | SiMYB16 | MYB-related | CSE, FAR1, CYP87A3, 4CL1, PAL, F5H, COMT, NCED3 | 调控植物木质素、类黄酮及木栓质生物合成 | 盐分 | [ |
| 木薯 Manihot esculenta | MeMYB60 | R2R3-MYB | CAT1/2 | 参与保卫细胞中活性氧稳态并影响气孔运动 | 脱落酸、干旱 | [ |
| 葡萄 Vitis amurensis | VaMYB14 | R2R3-MYB | ABA signaling genes, CORs, LTPs, CAT, POD | 参与激活ABA信号组分和CBF-COR非依赖的LTP3表达 | 盐分、低温、干旱 | [ |
| 番茄 Solanum lycopersicum | SlMYB41 | R2R3-MYB | SlHSP90.3 | 维持热胁迫下的活性氧稳态 | 高温 | [ |
| [1] | Li JG, Pu LJ, Han MF, et al. Soil salinization research in China: Advances and prospects[J]. Journal of Geographical Sciences, 2014, 24(5): 943-960. |
| [2] |
邱文怡, 王诗雨, 李晓芳, 等. MYB转录因子参与植物非生物胁迫响应与植物激素应答的研究进展[J]. 浙江农业学报, 2020, 32(7): 1317-1328.
doi: 10.3969/j.issn.1004-1524.2020.07.21 |
|
Qiu WY, Wang SY, Li XF, et al. Functions of plant MYB transcription factors in response to abiotic stress and plant hormones[J]. Acta Agric Zhejiangensis, 2020, 32(7): 1317-1328.
doi: 10.3969/j.issn.1004-1524.2020.07.21 |
|
| [3] | Sun JH, Xu JD, Qu WR, et al. Genome-wide analysis of R2R3-MYB transcription factors reveals their differential responses to drought stress and ABA treatment in desert poplar(Populus euphratica)[J]. Gene, 2023, 855: 147124. |
| [4] | Li XX, Guo C, Li ZY, et al. Deciphering the roles of tobacco MYB transcription factors in environmental stress tolerance[J]. Front Plant Sci, 2022, 13: 998606. |
| [5] | Yang JH, Zhang BH, Gu G, et al. Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco(Nicotiana tabacum L.)[J]. BMC Genomics, 2022, 23(1): 432. |
| [6] |
Paz-Ares J, Ghosal D, Wienand U, et al. The regulatory c1 locus of Zea mays encodes a protein with homology to MYB proto-oncogene products and with structural similarities to transcriptional activators[J]. EMBO J, 1987, 6(12): 3553-3558.
doi: 10.1002/j.1460-2075.1987.tb02684.x pmid: 3428265 |
| [7] | Chen YH, Yang XY, He K, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family[J]. Plant Mol Biol, 2006, 60(1): 107-124. |
| [8] | Kang LH, Teng YY, Cen QW, et al. Genome-wide identification of R2R3-MYB transcription factor and expression analysis under abiotic stress in rice[J]. Plants, 2022, 11(15): 1928. |
| [9] |
Du H, Yang SS, Liang Z, et al. Genome-wide analysis of the MYB transcription factor superfamily in soybean[J]. BMC Plant Biol, 2012, 12: 106.
doi: 10.1186/1471-2229-12-106 pmid: 22776508 |
| [10] |
Stracke R, Holtgräwe D, Schneider J, et al. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet(Beta vulgaris)[J]. BMC Plant Biol, 2014, 14: 249.
doi: 10.1186/s12870-014-0249-8 pmid: 25249410 |
| [11] | Wu Y, Wen J, Xia YP, et al. Evolution and functional diversification of R2R3-MYB transcription factors in plants[J]. Hortic Res, 2022, 9: uhac058. |
| [12] | Li ZJ, Peng RH, Tian YS, et al. Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum[J]. Plant Cell Physiol, 2016, 57(8): 1657-1677. |
| [13] | Chen GQ, He WZ, Guo XX, et al. Genome-wide identification, classification and expression analysis of the MYB transcription factor family in Petunia[J]. Int J Mol Sci, 2021, 22(9): 4838. |
| [14] | Xia H, Liu XL, Lin ZY, et al. Genome-wide identification of MYB transcription factors and screening of members involved in stress response in Actinidia[J]. Int J Mol Sci, 2022, 23(4): 2323. |
| [15] | Arce-Rodríguez ML, Martínez O, Ochoa-Alejo N. Genome-wide identification and analysis of the MYB transcription factor gene family in chili pepper(Capsicum spp.)[J]. Int J Mol Sci, 2021, 22(5): 2229. |
| [16] | Tan L, Ijaz U, Salih H, et al. Genome-wide identification and comparative analysis of MYB transcription factor family in Musa acuminata and Musa balbisiana[J]. Plants, 2020, 9(4): 413. |
| [17] | Xie FF, Hua QZ, Chen CB, et al. Genome-wide characterization of R2R3-MYB transcription factors in pitaya reveals a R2R3-MYB repressor HuMYB1 involved in fruit ripening through regulation of betalain biosynthesis by repressing betalain biosynthesis-related genes[J]. Cells, 2021, 10(8): 1949. |
| [18] | Li YM, Liang J, Zeng XZ, et al. Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis[J]. Hortic Plant J, 2021, 7(2): 129-141. |
| [19] | Muleke EM, Wang Y, Zhang WT, et al. Genome-wide identification and expression profiling of MYB transcription factor genes in radish(Raphanus sativus L.)[J]. J Integr Agric, 2021, 20(1): 120-131. |
| [20] | He CM, Teixeira da Silva JA, Wang HB, et al. Mining MYB transcription factors from the genomes of orchids(Phalaenopsis and Dendrobium)and characterization of an orchid R2R3-MYB gene involved in water-soluble polysaccharide biosynthesis[J]. Sci Rep, 2019, 9(1): 13818. |
| [21] |
Bertioli DJ, Jenkins J, Clevenger J, et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea[J]. Nat Genet, 2019, 51(5): 877-884.
doi: 10.1038/s41588-019-0405-z pmid: 31043755 |
| [22] | Chen QC, Zhang XD, Fang YX, et al. Genome-wide identification and expression analysis of the R2R3-MYB transcription factor family revealed their potential roles in the flowering process in longan(Dimocarpus longan)[J]. Front Plant Sci, 2022, 13: 820439. |
| [23] | Li PF, Wen J, Chen P, et al. MYB superfamily in Brassica napus: evidence for hormone-mediated expression profiles, large expansion, and functions in root hair development[J]. Biomolecules, 2020, 10(6): 875. |
| [24] | Wang YJ, Zhang Y, Fan CJ, et al. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia[J]. BMC Plant Biol, 2021, 21(1): 328. |
| [25] | Sabir IA, Manzoor MA, Shah IH, et al. MYB transcription factor family in sweet cherry(Prunus avium L.): genome-wide investigation, evolution, structure, characterization and expression patterns[J]. BMC Plant Biol, 2022, 22(1): 2. |
| [26] | Chen Z, Wu ZX, Dong WY, et al. MYB transcription factors becoming mainstream in plant roots[J]. Int J Mol Sci, 2022, 23(16): 9262. |
| [27] |
Wang L, Qiu TQ, Yue JR, et al. Arabidopsis ADF1 is regulated by MYB73 and is involved in response to salt stress affecting actin filament organization[J]. Plant Cell Physiol, 2021, 62(9): 1387-1395.
doi: 10.1093/pcp/pcab081 pmid: 34086948 |
| [28] |
He J, Liu YQ, Yuan DY, et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice[J]. Proc Natl Acad Sci USA, 2020, 117(1): 271-277.
doi: 10.1073/pnas.1902771116 pmid: 31848246 |
| [29] | Lu HC, Lam SH, Zhang DY, et al. R2R3-MYB genes coordinate conical cell development and cuticular wax biosynthesis in Phalaenopsis aphrodite[J]. Plant Physiol, 2022, 188(1): 318-331. |
| [30] | Yang Y, Zhang LB, Chen P, et al. UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development[J]. EMBO J, 2020, 39(2): e101928. |
| [31] |
Ma DW, Constabel CP. MYB repressors as regulators of phenylpropanoid metabolism in plants[J]. Trends Plant Sci, 2019, 24(3): 275-289.
doi: S1360-1385(18)30290-5 pmid: 30704824 |
| [32] | Zheng T, Tan WR, Yang H, et al. Regulation of anthocyanin accumulation via MYB75/HAT1/TPL-mediated transcriptional repression[J]. PLoS Genet, 2019, 15(3): e1007993. |
| [33] | Wang YM, Wang C, Guo HY, et al. BplMYB46 from Betula platyphylla can form homodimers and heterodimers and is involved in salt and osmotic stresses[J]. Int J Mol Sci, 2019, 20(5): 1171. |
| [34] | Shi MY, Liu X, Zhang HP, et al. The IAA- and ABA-responsive transcription factor CgMYB58 upregulates lignin biosynthesis and triggers juice sac granulation in pummelo[J]. Hortic Res, 2020, 7: 139. |
| [35] |
Wei XP, Lu WJ, Mao LC, et al. ABF2 and MYB transcription factors regulate feruloyl transferase FHT involved in ABA-mediated wound suberization of kiwifruit[J]. J Exp Bot, 2020, 71(1): 305-317.
doi: 10.1093/jxb/erz430 pmid: 31559426 |
| [36] | Sun WJ, Ma ZT, Chen H, et al. MYB gene family in potato(Solan-um tuberosum L.): genome-wide identification of hormone-responsive reveals their potential functions in growth and development[J]. Int J Mol Sci, 2019, 20(19): 4847. |
| [37] | Zhang S, Zhao QC, Zeng DX, et al. RhMYB108, an R2R3-MYB transcription factor, is involved in ethylene- and JA-induced petal senescence in rose plants[J]. Hortic Res, 2019, 6: 131. |
| [38] | Zhang L, Xu Y, Li YT, et al. Transcription factor CsMYB77 negatively regulates fruit ripening and fruit size in citrus[J]. Plant Physiol, 2024, 194(2): 867-883. |
| [39] | Wang XP, Niu YL, Zheng Y. Multiple functions of MYB transcription factors in abiotic stress responses[J]. Int J Mol Sci, 2021, 22(11): 6125. |
| [40] | Chen ZH, Teng SZ, Liu D, et al. RLM1, encoding an R2R3 MYB transcription factor, regulates the development of secondary cell wall in rice[J]. Front Plant Sci, 2022, 13: 905111. |
| [41] |
Pratyusha DS, Sarada DVL. MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses[J]. Plant Cell Rep, 2022, 41(12): 2245-2260.
doi: 10.1007/s00299-022-02927-1 pmid: 36171500 |
| [42] |
Gong ZZ. Plant abiotic stress: new insights into the factors that activate and modulate plant responses[J]. J Integr Plant Biol, 2021, 63(3): 429-430.
doi: 10.1111/jipb.13079 |
| [43] | Beathard C, Mooney S, Al-Saharin R, et al. Characterization of Arabidopsis thaliana R2R3 S23 MYB transcription factors as novel targets of the ubiquitin proteasome-pathway and regulators of salt stress and abscisic acid response[J]. Front Plant Sci, 2021, 12: 629208. |
| [44] | Li YY, Tian B, Wang Y, et al. The transcription factor MYB37 positively regulates photosynthetic inhibition and oxidative damage in Arabidopsis leaves under salt stress[J]. Front Plant Sci, 2022, 13: 943153. |
| [45] | Li X, Lu JH, Zhu XL, et al. AtMYBS1 negatively regulates heat tolerance by directly repressing the expression of MAX1 required for strigolactone biosynthesis in Arabidopsis[J]. Plant Commun, 2023, 4(6): 100675. |
| [46] | Ortiz-García P, Pérez-Alonso MM, González Ortega-Villaizán A, et al. The indole-3-acetamide-induced Arabidopsis transcription factor MYB74 decreases plant growth and contributes to the control of osmotic stress responses[J]. Front Plant Sci, 2022, 13: 928386. |
| [47] | Wang FB, Kong WL, Wong G, et al. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana[J]. Mol Genet Genomics, 2016, 291(4): 1545-1559. |
| [48] | Cheng YX, Ma YX, Zhang N, et al. The R2R3 MYB transcription factor MYB71 regulates abscisic acid response in Arabidopsis[J]. Plants, 2022, 11(10): 1369. |
| [49] | Lee SB, Kim HU, Suh MC. MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis[J]. Plant Cell Physiol, 2016, 57(11): 2300-2311. |
| [50] | Chen N, Pan LJ, Yang Z, et al. A MYB-related transcription factor from peanut, AhMYB30, improves freezing and salt stress tolerance in transgenic Arabidopsis through both DREB/CBF and ABA-signaling pathways[J]. Front Plant Sci, 2023, 14: 1136626. |
| [51] | Chen XS, Wu Y, Yu ZH, et al. BcMYB111 responds to BcCBF2 and induces flavonol biosynthesis to enhance tolerance under cold stress in non-heading Chinese cabbage[J]. Int J Mol Sci, 2023, 24(10): 8670. |
| [52] | Yang LJ, Chen Y, Xu L, et al. The OsFTIP6-OsHB22-OsMYBR57 module regulates drought response in rice[J]. Mol Plant, 2022, 15(7): 1227-1242. |
| [53] | Tiwari P, Indoliya Y, Chauhan AS, et al. Over-expression of rice R1-type MYB transcription factor confers different abiotic stress tolerance in transgenic Arabidopsis[J]. Ecotoxicol Environ Saf, 2020, 206: 111361. |
| [54] | Qu XX, Zou JJ, Wang JX, et al. A rice R2R3-type MYB transcription factor OsFLP positively regulates drought stress response via OsNAC[J]. Int J Mol Sci, 2022, 23(11): 5873. |
| [55] | Liu TL, Chen TZ, Kan JL, et al. The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants[J]. Plant Biotechnol J, 2022, 20(4): 722-735. |
| [56] | Liu RD, Shen YH, Wang MX, et al. GhMYB102 promotes drought resistance by regulating drought-responsive genes and ABA biosynthesis in cotton(Gossypium hirsutum L.)[J]. Plant Sci, 2023, 329: 111608. |
| [57] | Han GL, Qiao ZQ, Li YX, et al. LbMYB48 positively regulates salt gland development of Limonium bicolor and salt tolerance of plants[J]. Front Plant Sci, 2022, 13: 1039984. |
| [58] | Leng BY, Wang X, Yuan F, et al. Heterologous expression of the Limonium bicolor MYB transcription factor LbTRY in Arabidopsis thaliana increases salt sensitivity by modifying root hair development and osmotic homeostasis[J]. Plant Sci, 2021, 302: 110704. |
| [59] | Wang YG, Ye HY, Wang KT, et al. CcMYB12 positively regulates flavonoid accumulation during fruit development in Carya cathayensis and has a role in abiotic stress responses[J]. Int J Mol Sci, 2022, 23(24): 15618. |
| [60] | Qin B, Fan SL, Yu HY, et al. HbMYB44, a rubber tree MYB transcription factor with versatile functions in modulating multiple phytohormone signaling and abiotic stress responses[J]. Front Plant Sci, 2022, 13: 893896. |
| [61] | Wang C, Wang LJ, Lei J, et al. IbMYB308, a sweet potato R2R3-MYB gene, improves salt stress tolerance in transgenic tobacco[J]. Genes, 2022, 13(8): 1476. |
| [62] | Wang Z, Li X, Gao XR, et al. IbMYB73 targets abscisic acid-responsive IbGER5 to regulate root growth and stress tolerance in sweet potato[J]. Plant Physiol, 2024, 194(2): 787-804. |
| [63] | Zhou ZX, Wei XX, Lan HY. CgMYB1, an R2R3-MYB transcription factor, can alleviate abiotic stress in an annual halophyte Chenopodium glaucum[J]. Plant Physiol Biochem, 2023, 196: 484-496. |
| [64] | Ning CH, Yang YT, Chen QY, et al. An R2R3 MYB transcription factor PsFLP regulates the symmetric division of guard mother cells during stomatal development in Pisum sativum[J]. Physiol Plant, 2023, 175(3): e13943. |
| [65] | Zhou LD, Huan XH, Zhao K, et al. PagMYB205 negatively affects poplar salt tolerance through reactive oxygen species scavenging and root vitality modulation[J]. Int J Mol Sci, 2023, 24(20): 15437. |
| [66] | Hu J, Zou SQ, Huang JJ, et al. PagMYB151 facilitates proline accumulation to enhance salt tolerance of poplar[J]. BMC Genomics, 2023, 24(1): 345. |
| [67] | Ren CK, Li ZH, Song PH, et al. Overexpression of a grape MYB transcription factor gene VhMYB2 increases salinity and drought tolerance in Arabidopsis thaliana[J]. Int J Mol Sci, 2023, 24(13): 10743. |
| [68] | Lim J, Lim CW, Lee SC. Role of pepper MYB transcription factor CaDIM1 in regulation of the drought response[J]. Front Plant Sci, 2022, 13: 1028392. |
| [69] | Chen Q, Peng L, Wang AH, et al. An R2R3-MYB FtMYB11 from Tartary buckwheat has contrasting effects on abiotic tolerance in Arabidopsis[J]. J Plant Physiol, 2023, 280: 153842. |
| [70] | Sun M, Xu QY, Zhu ZP, et al. AgMYB5, an MYB transcription factor from celery, enhanced β-carotene synthesis and promoted drought tolerance in transgenic Arabidopsis[J]. BMC Plant Biol, 2023, 23(1): 151. |
| [71] | Yu Y, Guo DD, Min DH, et al. Foxtail millet MYB-like transcription factor SiMYB16 confers salt tolerance in transgenic rice by regulating phenylpropane pathway[J]. Plant Physiol Biochem, 2023, 195: 310-321. |
| [72] |
徐子寅, 于晓玲, 邹良平, 等. 木薯MYB转录因子基因MeMYB60表达特征分析及其互作蛋白筛选[J]. 作物学报, 2023, 49(4): 955-965.
doi: 10.3724/SP.J.1006.2023.24089 |
| Xu ZY, Yu XL, Zou LP, et al. Expression pattern analysis and interaction protein screening of cassava MYB transcription factor MeMYB60[J]. Acta Agron Sin, 2023, 49(4): 955-965. | |
| [73] | Fang LC, Wang ZM, Su LY, et al. Vitis MYB14 confer cold and drought tolerance by activating lipid transfer protein genes expression and reactive oxygen species scavenge[J]. Gene, 2024, 890: 147792. |
| [74] | Wang JY, Chen C, Wu CZ, et al. SlMYB41 positively regulates tomato thermotolerance by activating the expression of SlHSP90.3[J]. Plant Physiol Biochem, 2023, 204: 108106. |
| [75] | Zhao HX, Yao PF, Zhao JL, et al. A novel R2R3-MYB transcription factor FtMYB22 negatively regulates salt and drought stress through ABA-dependent pathway[J]. Int J Mol Sci, 2022, 23(23): 14549. |
| [76] | Wang S, Wu HL, Cao XX, et al. Tartary buckwheat FtMYB30 transcription factor improves the salt/drought tolerance of transgenic Arabidopsis in an ABA-dependent manner[J]. Physiol Plant, 2022, 174(5): e13781. |
| [77] | Chen C, Zhang Z, Lei YY, et al. MdMYB44-like positively regulates salt and drought tolerance via the MdPYL8-MdPP2CA module in apple[J]. Plant J, 2023: 38102874. DOI: 10.1111/tpj.16584. |
| [78] |
Chen KQ, Song MR, Guo YN, et al. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals[J]. Plant Biotechnol J, 2019, 17(12): 2341-2355.
doi: 10.1111/pbi.13151 pmid: 31077628 |
| [79] | Zhang P, Wang RL, Yang XP, et al. The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence[J]. Plant Cell Environ, 2020, 43(8): 1925-1943. |
| [80] | Shukla V, Han JP, Cléard F, et al. Suberin plasticity to developmental and exogenous cues is regulated by a set of MYB transcription factors[J]. Proc Natl Acad Sci USA, 2021, 118(39): e2101730118. |
| [81] | Kim D, Jeon SJ, Yanders S, et al. MYB3 plays an important role in lignin and anthocyanin biosynthesis under salt stress condition in Arabidopsis[J]. Plant Cell Rep, 2022, 41(7): 1549-1560. |
| [82] |
Adler G, Blumwald E, Bar-Zvi D. The sugar beet gene encoding the sodium/proton exchanger 1(BvNHX1)is regulated by a MYB transcription factor[J]. Planta, 2010, 232(1): 187-195.
doi: 10.1007/s00425-010-1160-7 pmid: 20390294 |
| [83] | Zhang X, Chen LC, Shi QH, et al. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato[J]. Plant Sci, 2020, 291: 110356. |
| [84] | Sun YH, Zhao J, Li XY, et al. E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in Arabidopsis[J]. New Phytol, 2020, 227(2): 455-472. |
| [85] | Zhu ZG, Quan R, Chen GX, et al. An R2R3-MYB transcription factor VyMYB24, isolated from wild grape Vitis yanshanesis J. X. Chen., regulates the plant development and confers the tolerance to drought[J]. Front Plant Sci, 2022, 13: 966641. |
| [86] | Wu JD, Jiang YL, Liang YN, et al. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants[J]. Plant Physiol Biochem, 2019, 137: 179-188. |
| [87] | Li HQ, Yang J, Ma RJ, et al. Genome-wide identification and expression analysis of MYB gene family in Cajanus cajan and CcMYB107 improves plant drought tolerance[J]. Physiol Plant, 2023, 175(4): e13954. |
| [88] | Dong NQ, Lin HX. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions[J]. J Integr Plant Biol, 2021, 63(1): 180-209. |
| [89] | Ren YS, Zhang SY, Zhao QY, et al. The CsMYB123 and CsbHLH111 are involved in drought stress-induced anthocyanin biosynthesis in Chaenomeles speciosa[J]. Mol Hortic, 2023, 3(1): 25. |
| [90] | Chen YN, Feng PP, Zhang XW, et al. Silencing of SlMYB50 affects tolerance to drought and salt stress in tomato[J]. Plant Physiol Biochem, 2022, 193: 139-152. |
| [91] | Chen YN, Li L, Tang BY, et al. Silencing of SlMYB55 affects plant flowering and enhances tolerance to drought and salt stress in tomato[J]. Plant Sci, 2022, 316: 111166. |
| [92] | Zhu N, Duan BL, Zheng HL, et al. An R2R3 MYB gene GhMYB3 functions in drought stress by negatively regulating stomata movement and ROS accumulation[J]. Plant Physiol Biochem, 2023, 197: 107648. |
| [93] | Zhao HY, Zhao HQ, Hu YF, et al. Expression of the sweet potato MYB transcription factor IbMYB48 confers salt and drought tolerance in Arabidopsis[J]. Genes, 2022, 13(10): 1883. |
| [94] | Liu YH, Shen Y, Liang M, et al. Identification of peanut AhMYB44 transcription factors and their multiple roles in drought stress responses[J]. Plants, 2022, 11(24): 3522. |
| [95] |
Gupta A, Rico-Medina A, Caño-Delgado AI. The physiology of plant responses to drought[J]. Science, 2020, 368(6488): 266-269.
doi: 10.1126/science.aaz7614 pmid: 32299946 |
| [96] |
Gao J, Zhao Y, Zhao ZK, et al. RRS1 shapes robust root system to enhance drought resistance in rice[J]. New Phytol, 2023, 238(3): 1146-1162.
doi: 10.1111/nph.18775 pmid: 36862074 |
| [97] | Song Q, Kong LF, Yang XR, et al. PtoMYB142, a poplar R2R3-MYB transcription factor, contributes to drought tolerance by regulating wax biosynthesis[J]. Tree Physiol, 2022, 42(10): 2133-2147. |
| [98] | Hsu PK, Dubeaux G, Takahashi Y, et al. Signaling mechanisms in abscisic acid-mediated stomatal closure[J]. Plant J, 2021, 105(2): 307-321. |
| [99] | Dossa K, Mmadi MA, Zhou R, et al. Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis[J]. AoB Plants, 2019, 12(1): plz081. |
| [100] | Duan BL, Xie XF, Jiang YH, et al. GhMYB44 enhances stomatal closure to confer drought stress tolerance in cotton and Arabidopsis[J]. Plant Physiol Biochem, 2023, 198: 107692. |
| [101] | Chen LM, Yang HL, Fang YS, et al. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway[J]. Plant Biotechnol J, 2021, 19(4): 702-716. |
| [102] |
Ding YL, Shi YT, Yang SH. Molecular regulation of plant responses to environmental temperatures[J]. Mol Plant, 2020, 13(4): 544-564.
doi: S1674-2052(20)30034-4 pmid: 32068158 |
| [103] | Abdullah SNA, Azzeme AM, Yousefi K. Fine-tuning cold stress response through regulated cellular abundance and mechanistic actions of transcription factors[J]. Front Plant Sci, 2022, 13: 850216. |
| [104] | Yao CY, Li XG, Li YM, et al. Overexpression of a Malus baccata MYB transcription factor gene MbMYB4 increases cold and drought tolerance in Arabidopsis thaliana[J]. Int J Mol Sci, 2022, 23(3): 1794. |
| [105] | Yao CY, Li WH, Liang XQ, et al. Molecular cloning and characterization of MbMYB108, a Malus baccata MYB transcription factor gene, with functions in tolerance to cold and drought stress in transgenic Arabidopsis thaliana[J]. Int J Mol Sci, 2022, 23(9): 4846. |
| [106] | Liu WD, Wang TH, Wang Y, et al. MbMYBC1, a M. baccata MYB transcription factor, contribute to cold and drought stress tolerance in transgenic Arabidopsis[J]. Front Plant Sci, 2023, 14: 1141446. |
| [107] | Luo D, Raza A, Cheng Y, et al. Cloning and functional characterization of cold-inducible MYB-like 17 transcription factor in rapeseed(Brassica napus L.)[J]. Int J Mol Sci, 2023, 24(11): 9514. |
| [108] | Dar NA, Mir MA, Mir JI, et al. MYB-6 and LDOX-1 regulated accretion of anthocyanin response to cold stress in purple black carrot(Daucus carota L.)[J]. Mol Biol Rep, 2022, 49(6): 5353-5364. |
| [109] | Zhang TT, Cui Z, Li YX, et al. Genome-wide identification and expression analysis of MYB transcription factor superfamily in Dendrobium catenatum[J]. Front Genet, 2021, 12: 714696. |
| [110] | Wu WH, Zhu S, Zhu LM, et al. Characterization of the Liriodendron chinense MYB gene family and its role in abiotic stress response[J]. Front Plant Sci, 2021, 12: 641280. |
| [111] | Lv JH, Xu Y, Dan XM, et al. Genomic survey of MYB gene family in six pearl millet(Pennisetum glaucum)varieties and their response to abiotic stresses[J]. Genetica, 2023, 151(3): 251-265. |
| [112] | Jacob P, Brisou G, Dalmais M, et al. The seed development factors TT2 and MYB5 regulate heat stress response in Arabidopsis[J]. Genes, 2021, 12(5): 746. |
| [113] | Wang HZ, Pak S, Yang J, et al. Two high hierarchical regulators, PuMYB40 and PuWRKY75, control the low phosphorus driven adventitious root formation in Populus ussuriensis[J]. Plant Biotechnol J, 2022, 20(8): 1561-1577. |
| [114] | Sega P, Pacak A. Plant PHR transcription factors: put on A map[J]. Genes, 2019, 10(12): 1018. |
| [115] | Li YJ, Ma WJ, Zhang KF, et al. Overexpression of GmPHR1 promotes soybean yield through global regulation of nutrient acquisition and root development[J]. Int J Mol Sci, 2022, 23(23): 15274. |
| [116] | Rerkasem B, Jamjod S, Pusadee T. Productivity limiting impacts of boron deficiency, a review[J]. Plant Soil, 2020, 455(1): 23-40. |
| [117] | Liu J, Chen T, Wang CL, et al. Transcriptome analysis in Pyrus betulaefolia roots in response to short-term boron deficiency[J]. Genes, 2023, 14(4): 817. |
| [118] |
Gao LJ, Liu XP, Gao KK, et al. ART1 and putrescine contribute to rice aluminum resistance via OsMYB30 in cell wall modification[J]. J Integr Plant Biol, 2023, 65(4): 934-949.
doi: 10.1111/jipb.13429 |
| [119] | Wu Q, Tao Y, Huang J, et al. The MYB transcription factor MYB103 acts upstream of TRICHOME BIREFRINGENCE-LIKE27 in regulating aluminum sensitivity by modulating the O-acetylation level of cell wall xyloglucan in Arabidopsis thaliana[J]. Plant J, 2022, 111(2): 529-545. |
| [120] | Wang HJ, Yin XL, Du D, et al. GsMYB7 encoding a R2R3-type MYB transcription factor enhances the tolerance to aluminum stress in soybean(Glycine max L.)[J]. BMC Genomics, 2022, 23(1): 529. |
| [121] | Feng SG, Hou KL, Zhang HS, et al. Investigation of the role of TmMYB16/123 and their targets(TmMTP1/11)in the tolerance of Taxus media to cadmium[J]. Tree Physiol, 2023, 43(6): 1009-1022. |
| [122] |
Sapara KK, Khedia J, Agarwal P, et al. SbMYB15 transcription factor mitigates cadmium and nickel stress in transgenic tobacco by limiting uptake and modulating antioxidative defence system[J]. Funct Plant Biol, 2019, 46(8): 702-714.
doi: 10.1071/FP18234 pmid: 31023418 |
| [123] | Su LT, Lv AM, Wen WW, et al. MsMYB741 is involved in alfalfa resistance to aluminum stress by regulating flavonoid biosynthesis[J]. Plant J, 2022, 112(3): 756-771. |
| [124] | Biswas D, Gain H, Mandal A. MYB transcription factor: a new weapon for biotic stress tolerance in plants[J]. Plant Stress, 2023, 10: 100252. |
| [125] | Yu YB, Zhang S, Yu Y, et al. The pivotal role of MYB transcription factors in plant disease resistance[J]. Planta, 2023, 258(1): 16. |
| [126] | Kim SH, Lam PY, Lee MH, et al. The Arabidopsis R2R3 MYB transcription factor MYB15 is a key regulator of lignin biosynthesis in effector-triggered immunity[J]. Front Plant Sci, 2020, 11: 583153. |
| [127] | An C, Sheng LP, Du XP, et al. Overexpression of CmMYB15 provides chrysanthemum resistance to aphids by regulating the biosynthesis of lignin[J]. Hortic Res, 2019, 6: 84. |
| [128] | Wang YJ, Sheng LP, Zhang HR, et al. CmMYB19 over-expression improves aphid tolerance in Chrysanthemum by promoting lignin synthesis[J]. Int J Mol Sci, 2017, 18(3): 619. |
| [129] | Zhu YT, Hu XQ, Wang P, et al. GhODO1, an R2R3-type MYB transcription factor, positively regulates cotton resistance to Verticillium dahliae via the lignin biosynthesis and jasmonic acid signaling pathway[J]. Int J Biol Macromol, 2022, 201: 580-591. |
| [130] | Castorina G, Bigelow M, Hattery T, et al. Roles of the MYB94/FUSED LEAVES1(ZmFDL1)and GLOSSY2(ZmGL2)genes in cuticle biosynthesis and potential impacts on Fusarium verticillioides growth on maize silks[J]. Front Plant Sci, 2023, 14: 1228394. |
| [131] | Xiao SH, Hu Q, Shen JL, et al. GhMYB4 downregulates lignin biosynthesis and enhances cotton resistance to Verticillium dahliae[J]. Plant Cell Rep, 2021, 40(4): 735-751. |
| [132] | Zheng HY, Dong LL, Han XY, et al. The TuMYB46L-TuACO3 module regulates ethylene biosynthesis in einkorn wheat defense to powdery mildew[J]. New Phytol, 2020, 225(6): 2526-2541. |
| [133] | Zhang MF, Wang JQ, Luo QJ, et al. CsMYB96 enhances citrus fruit resistance against fungal pathogen by activating salicylic acid biosynthesis and facilitating defense metabolite accumulation[J]. J Plant Physiol, 2021, 264: 153472. |
| [134] | Gu KD, Zhang QY, Yu JQ, et al. R2R3-MYB transcription factor MdMYB73 confers increased resistance to the fungal pathogen Botryosphaeria dothidea in apples via the salicylic acid pathway[J]. J Agric Food Chem, 2021, 69(1): 447-458. |
| [1] | QIN Jian, LI Zhen-yue, HE Lang, LI Jun-ling, ZHANG Hao, DU Rong. Change of Single-cell Transcription Profile and Analysis of Intercellular Communication in Myogenic Cell Differentiation [J]. Biotechnology Bulletin, 2024, 40(6): 330-342. |
| [2] | DU Bing-shuai, ZOU Xin-hui, WANG Zi-hao, ZHANG Xin-yuan, CAO Yi-bo, ZHANG Ling-yun. Genome-wide Identification and Expression Analysis of the SWEET Gene Family in Camellia oleifera [J]. Biotechnology Bulletin, 2024, 40(5): 179-190. |
| [3] | GUO Hui-yan, DONG Xue, AN Meng-nan, XIA Zi-hao, WU Yuan-hua. Research Progress in the Functions of Key Enzymes of Ubiquitination Modification in Plant Stress Responses [J]. Biotechnology Bulletin, 2024, 40(4): 1-11. |
| [4] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [5] | CHEN Qiang, HUANG Xin-hui, ZHANG Zheng, ZHANG Chong, LIU Ye-fei. Effects of Melatonin on the Fruit Softening and Ethylene Synthesis of Post-harvest Oriental Melon [J]. Biotechnology Bulletin, 2024, 40(4): 139-147. |
| [6] | ZHANG Yu, SHI Lei, GONG Lei, NIE Feng-jie, YANG Jiang-wei, LIU Xuan, YANG Wen-jing, ZHANG Guo-hui, XIE Rui-xia, ZHANG Li. Genome-wide Identification of Potato WOX Gene Family and Its Expression Analysis in in vitro Regeneration and Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 170-180. |
| [7] | WU Xing-xing, HONG Hai-bo, GAN Zhi-cheng, LI Rui-ning, HUANG Xian-zhong. Cloning and Preliminary Functional Analysis of CaPI Gene in Capsicum annuum L. [J]. Biotechnology Bulletin, 2024, 40(3): 193-201. |
| [8] | JIANG Lin-qi, ZHAO Jia-ying, ZHENG Fei-xiong, YAO Xin-yi, LI Xiao-xian, YU Zhen-ming. Identification and Expression Analysis of 14-3-3 Gene Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2024, 40(3): 229-241. |
| [9] | ZHOU Hong-dan, LUO Xiao-ping, TU Mi-xue, LI Zhong-guang. Phytomelatonin: An Emerging Signal Molecule Responding to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 41-51. |
| [10] | WU Cui-cui, XIAO Shui-ping. Genome-wide Identification of HD-Zip Gene Family in Gossypium hirsutum L. and Expression Analysis in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(2): 130-145. |
| [11] | XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane [J]. Biotechnology Bulletin, 2024, 40(2): 197-211. |
| [12] | LI Ya-nan, ZHANG Hao-jie, LIANG Meng-jing, LUO Tao, LI Wang-ning, ZHANG Chun-hui, JI Chun-li, LI Run-zhi, XUE Jin-ai, CUI Hong-li. Identification and Expression Analysis of Calcium-dependent Protein Kinase(CDPK)Family in Haematococcus pluvialis [J]. Biotechnology Bulletin, 2024, 40(2): 300-312. |
| [13] | ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses [J]. Biotechnology Bulletin, 2024, 40(2): 90-98. |
| [14] | ZHANG Yi, ZHANG Xin-ru, ZHANG Jin-ke, HU Li-zong, SHANGGUAN Xin-xin, ZHENG Xiao-hong, HU Juan-juan, ZHANG Cong-cong, MU Gui-qing, LI Cheng-wei. Functional Analysis of TaMYB1 Gene in Wheat Under Cadmium Stress [J]. Biotechnology Bulletin, 2024, 40(1): 194-206. |
| [15] | WU Zhen, ZHANG Ming-Ying, YAN Feng, LI Yi-min, GAO Jing, YAN Yong-Gang, ZHANG Gang. Identification and Analysis of WRKY Gene Family in Rheum palmatum L. [J]. Biotechnology Bulletin, 2024, 40(1): 250-261. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||