








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (10): 210-221.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0473
Previous Articles Next Articles
LI Zhan-qian1( ), LI Chen1, LI Shu-ting1, MA Ju-hua1, JING Hai-qing1, SUN Yan1, ZHOU Ya-li1, XUE Jin-ai1,2(
), LI Chen1, LI Shu-ting1, MA Ju-hua1, JING Hai-qing1, SUN Yan1, ZHOU Ya-li1, XUE Jin-ai1,2( ), LI Run-zhi1,2(
), LI Run-zhi1,2( )
)
Received:2025-05-08
Online:2025-10-26
Published:2025-10-28
Contact:
XUE Jin-ai, LI Run-zhi
E-mail:lizhanqian2022@163.com;306214803@qq.com;rli2001@126.com
LI Zhan-qian, LI Chen, LI Shu-ting, MA Ju-hua, JING Hai-qing, SUN Yan, ZHOU Ya-li, XUE Jin-ai, LI Run-zhi. Genome-wide Identification of the ME Family in Cyperus esculentusis and Functional Analysis of CeNAD-ME2[J]. Biotechnology Bulletin, 2025, 41(10): 210-221.
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| CeNADP-ME1 | GAGGAGCTGTTGCCATCTGT | TGTGACAGATGGCGAACGG |
| CeNADP-ME2 | CACTGACGGAGGACGGATT | TCTGCGTGCTTGCCTATTACA |
| CeNADP-ME3 | GCATGAGAAGAGCATCCAGG | CTTCTGCGTGCCTGCCTATTA |
| CeNADP-ME4 | CTTCTGCCTCCTGCTATCGTT | TGGTTCCATCTTCAGGCGTC |
| CeNAD-ME1 | TGGTGACAGATGGAAGCAGGA | CTGTATGTCTCCGCTGCTGG |
| CeNAD-ME2 | AGAATGCTCGGCACTGACC | AGTGGTCAAGAAGGTGAAGCC |
| CeActin | TCGGTGGTATTGGAACTG | AAGCACTGGAGCGTAGCC |
| pBI121-CeNAD-ME2 | aacctgcaggtcgac | ggtttaaacgagctc |
| pYES2.0-CeNAD-ME2 | attaagcttggtacc | tacatgatgcggccc |
| 1300-CeNAD-ME2 | atacaccaaatcgac | cgatcggggaaattc |
Table 1 Primer sequences
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| CeNADP-ME1 | GAGGAGCTGTTGCCATCTGT | TGTGACAGATGGCGAACGG |
| CeNADP-ME2 | CACTGACGGAGGACGGATT | TCTGCGTGCTTGCCTATTACA |
| CeNADP-ME3 | GCATGAGAAGAGCATCCAGG | CTTCTGCGTGCCTGCCTATTA |
| CeNADP-ME4 | CTTCTGCCTCCTGCTATCGTT | TGGTTCCATCTTCAGGCGTC |
| CeNAD-ME1 | TGGTGACAGATGGAAGCAGGA | CTGTATGTCTCCGCTGCTGG |
| CeNAD-ME2 | AGAATGCTCGGCACTGACC | AGTGGTCAAGAAGGTGAAGCC |
| CeActin | TCGGTGGTATTGGAACTG | AAGCACTGGAGCGTAGCC |
| pBI121-CeNAD-ME2 | aacctgcaggtcgac | ggtttaaacgagctc |
| pYES2.0-CeNAD-ME2 | attaagcttggtacc | tacatgatgcggccc |
| 1300-CeNAD-ME2 | atacaccaaatcgac | cgatcggggaaattc |

Fig. 1 Amino acid sequence alignment of the CeME family proteins (A, B) and the evolutionary tree analysis (C) of CeMEs with ME proteins from other plantsI‒V: Five conserved domains of malic enzyme. Ce: Cyperus esculentusis; At: Arabidopsis thaliana; Zm: Zea mays (maize). Ta: Triticum aestivum (wheat); Os: Oryza sativa (rice); St: Solanum tuberosum (potato); Gm: Glycine max (soybean)
基因 Gene | 基因 ID Gene ID | 氨基酸数 Number of amino acids | 相对分子质量 Molecular weight (kD) | 理论等电点 pI | 不稳定系数 Instability factor | 亲水性 Hydrophilicity | 亚细胞定位预测 Prediction of subcellu-lar location |
|---|---|---|---|---|---|---|---|
| CeNADP-ME1 | CESC_03761 | 650 | 71.11 | 7.00 | 37.79 | -0.206 | 叶绿体 |
| CeNADP-ME2 | CESC_08481 | 577 | 64.01 | 5.97 | 41.44 | -0.112 | 叶绿体 |
| CeNADP-ME3 | CESC_16067 | 641 | 71.43 | 6.73 | 44.48 | -0.153 | 叶绿体 |
| CeNADP-ME4 | CESC_16827 | 662 | 73.62 | 6.78 | 49.33 | -0.121 | 叶绿体 |
| CeNAD-ME1 | CESC_15235 | 623 | 68.87 | 5.59 | 43.08 | -0.112 | 线粒体 |
| CeNAD-ME2 | CESC_17435 | 609 | 67.51 | 7.98 | 36.58 | -0.119 | 线粒体 |
Table 2 Analysis of the physiochemical properties of the ME family proteins in C. yperus esculentusis L.
基因 Gene | 基因 ID Gene ID | 氨基酸数 Number of amino acids | 相对分子质量 Molecular weight (kD) | 理论等电点 pI | 不稳定系数 Instability factor | 亲水性 Hydrophilicity | 亚细胞定位预测 Prediction of subcellu-lar location |
|---|---|---|---|---|---|---|---|
| CeNADP-ME1 | CESC_03761 | 650 | 71.11 | 7.00 | 37.79 | -0.206 | 叶绿体 |
| CeNADP-ME2 | CESC_08481 | 577 | 64.01 | 5.97 | 41.44 | -0.112 | 叶绿体 |
| CeNADP-ME3 | CESC_16067 | 641 | 71.43 | 6.73 | 44.48 | -0.153 | 叶绿体 |
| CeNADP-ME4 | CESC_16827 | 662 | 73.62 | 6.78 | 49.33 | -0.121 | 叶绿体 |
| CeNAD-ME1 | CESC_15235 | 623 | 68.87 | 5.59 | 43.08 | -0.112 | 线粒体 |
| CeNAD-ME2 | CESC_17435 | 609 | 67.51 | 7.98 | 36.58 | -0.119 | 线粒体 |

Fig. 7 Quantitative analysis of ME gene expression in different stages of C. esculentusis tuber developmentDifferent letters indicate significant differences at P<0.05. The same below
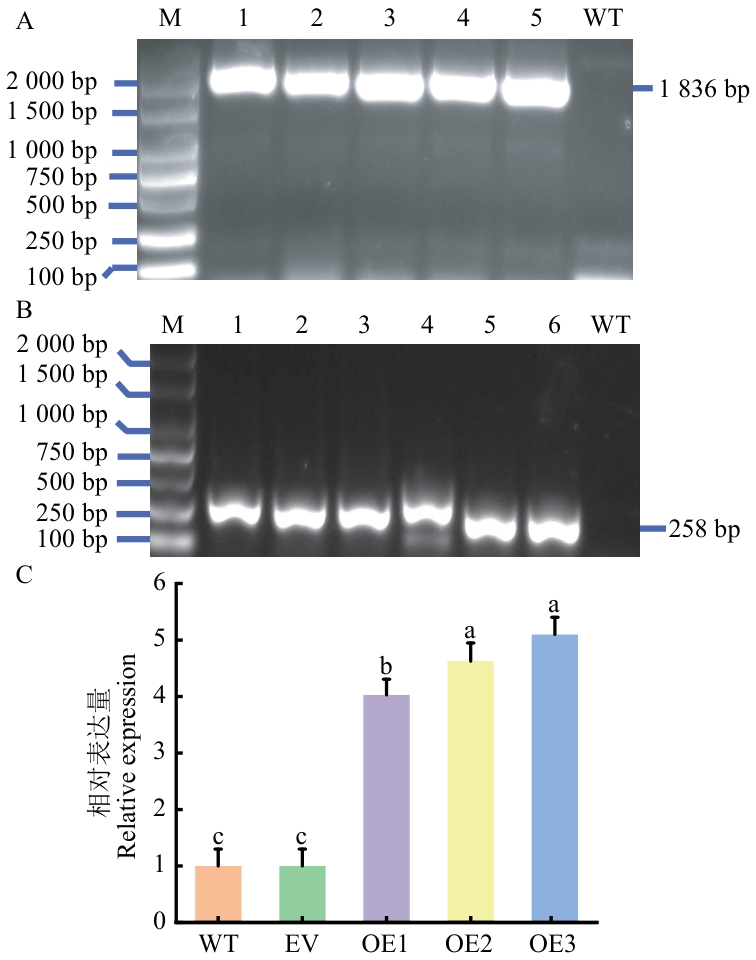
Fig. 11 DNA molecular identification (A), RNA molecule identification (B) and relative expression (C) of CeNAD-ME2-transgenic tobacco plantsM: DL2000 marker; 1‒6: transfer PBI121-CeNAD-ME2 positive band; WT: wild control; EV: empty vector control; OE: transgenic PBI121-CeNAD-ME2 positive plants. The same below
| [1] | Sánchez-Zapata E, Fernández-López J, Angel Pérez-Alvarez J. Tiger nut (Cyperus esculentus) commercialization: health aspects, composition, properties, and food applications [J]. Compr Rev Food Sci Food Saf, 2012, 11(4): 366-377. |
| [2] | Bullen JJ, Rogers HJ, Spalding PB, et al. Natural resistance, iron and infection: a challenge for clinical medicine [J]. J Med Microbiol, 2006, 55(Pt 3): 251-258. |
| [3] | 瞿萍梅, 程治英, 龙春林, 等. 油莎豆资源的综合开发利用 [J]. 中国油脂, 2007, 32(9): 61-63. |
| Qu PM, Cheng ZY, Long CL, et al. Comprehensive development of chufa (Cyperus esculentus L. var. sativus) [J]. China Oils Fats, 2007, 32(9): 61-63. | |
| [4] | Makareviciene V, Gumbyte M, Yunik A, et al. Opportunities for the use of chufa sedge in biodiesel production [J]. Ind Crops Prod, 2013, 50: 633-637. |
| [5] | Li T, Sun Y, Chen Y, et al. Characterisation of two novel genes encoding Δ9 fatty acid desaturases (CeSADs) for oleic acid accumulation in the oil-rich tuber of Cyperus esculentus [J]. Plant Sci, 2022, 319: 111243. |
| [6] | McCullough W, Roberts CF. The role of malic enzyme in Aspergillus nidulans [J]. FEBS Lett, 1974, 41(2): 238-242. |
| [7] | Boles E, de Jong-Gubbels P, Pronk JT. Identification and characterization of MAE1 the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme [J]. J Bacteriol, 1998, 180(11): 2875-2882. |
| [8] | Thelen JJ, Ohlrogge JB. Metabolic engineering of fatty acid biosynthesis in plants [J]. Metab Eng, 2002, 4(1): 12-21. |
| [9] | 周滈, 杨传平, 柳参奎. 植物苹果酸酶在抵御逆境中的作用 [J]. 林业科技, 2011, 36(6): 44-47. |
| Zhou H, Yang CP, Liu SK. The role of plant malic enzyme in defense against stress environments [J]. For Sci Technol, 2011, 36(6): 44-47. | |
| [10] | Rao XL, Dixon RA. The differences between NAD-ME and NADP-ME subtypes of C4 photosynthesis: more than decarboxylating enzymes [J]. Front Plant Sci, 2016, 7: 1525. |
| [11] | Wheeler MC, Tronconi MA, Drincovich MF, et al. A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis [J]. Plant Physiol, 2005, 139(1): 39-51. |
| [12] | Tronconi MA, Fahnenstich H, Gerrard Weehler MC, et al. Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism [J]. Plant Physiol, 2008, 146(4): 1540-1552. |
| [13] | Alvarez CE, Saigo M, Margarit E, et al. Kinetics and functional diversity among the five members of the NADP-malic enzyme family from Zea mays, a C4 species [J]. Photosynth Res, 2013, 115(1): 65-80. |
| [14] | Ratledge C. The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: a reappraisal and unsolved problems [J]. Biotechnol Lett, 2014, 36(8): 1557-1568. |
| [15] | Zhu BH, Zhang RH, Lv NN, et al. The role of malic enzyme on promoting total lipid and fatty acid production in Phaeodactylum tricornutum [J]. Front Plant Sci, 2018, 9: 826. |
| [16] | Zhang Y, Adams IP, Ratledge C. Malic enzyme: the controlling activity for lipid production overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation [J]. Microbiology, 2007, 153(Pt 7): 2013-2025. |
| [17] | Wynn JP, Ratledge C. Malic enzyme is a major source of NADPH for lipid accumulation by Aspergillus nidulans [J]. Microbiology, 1997, 143(1): 253-257. |
| [18] | Gerrard Wheeler MC, Arias CL, Righini S, et al. Differential contribution of malic enzymes during soybean and castor seeds maturation [J]. PLoS One, 2016, 11(6): e0158040. |
| [19] | Morley SA, Ma FF, Alazem M, et al. Expression of malic enzyme reveals subcellular carbon partitioning for storage reserve production in soybeans [J]. New Phytol, 2023, 239(5): 1834-1851. |
| [20] | Ji HY, Liu DT, Yang ZL. High oil accumulation in tuber of yellow nutsedge compared to purple nutsedge is associated with more abundant expression of genes involved in fatty acid synthesis and triacylglycerol storage [J]. Biotechnol Biofuels, 2021, 14(1): 54. |
| [21] | Gao Y, Sun Y, Gao HL, et al. Correction to: Ectopic overexpression of a type-II DGAT (CeDGAT2-2) derived from oil-rich tuber of Cyperus esculentus enhances accumulation of oil and oleic acid in tobacco leaves [J]. Biotechnol Biofuels, 2021, 14(1): 139. |
| [22] | 李秀峰, 张欣欣, 高野哲夫, 等. 水稻(Oryza sativa L.)苹果酸酶(OsNADP-ME3)基因在逆境下的表达特性研究 [J]. 基因组学与应用生物学, 2012, 31(4): 327-332. |
| Li XF, Zhang XX, Takano T, et al. Expression characteristics of rice (Oryza sativa L.) malic enzyme (OsNADP-ME3) gene under environmental stress [J]. Genom Appl Biol, 2012, 31(4): 327-332. | |
| [23] | 刘增辉. 小麦NADP-苹果酸酶与逆境关系的初步研究 [D]. 青岛: 青岛科技大学, 2010. |
| Liu ZH. Preliminary study on the relationg of NADP-malic enzyme and stresses [D]. Qingdao: Qingdao University of Science & Technology, 2010. | |
| [24] | Kawaoka A, Kawamoto T, Sekine M, et al. A cis-acting element and a trans-acting factor involved in the wound-induced expression of a horseradish peroxidase gene [J]. Plant J, 1994, 6(1): 87-97. |
| [25] | Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein [J]. Cell, 1998, 95(5): 657-667. |
| [26] | 张玉慧, 王玉兰, 夏春兰, 等. 松叶猪毛菜NADP-苹果酸酶基因家族及启动子克隆分析 [J]. 西北植物学报, 2022, 42(5): 760-769. |
| Zhang YH, Wang YL, Xia CL, et al. Cloning and analysis of NADP-ME gene family and promoters in Salsola laricifolia [J]. Acta Bot Boreali Occidentalia Sin, 2022, 42(5): 760-769. | |
| [27] | 史先飞, 高宇, 黄旭升, 等. 油莎豆丙酮酸激酶基因的鉴定及其在块茎发芽和幼苗形态建成时期的表达分析 [J]. 激光生物学报, 2022, 31(6): 533-541. |
| Shi XF, Gao Y, Huang XS, et al. Identification of pyruvate kinase genes and their expression analysis during Cyperus esculentus tuber germination and seedling establishment [J]. Acta Laser Biol Sin, 2022, 31(6): 533-541. | |
| [28] | Dao O, Kuhnert F, Weber APM, et al. Physiological functions of malate shuttles in plants and algae [J]. Trends Plant Sci, 2022, 27(5): 488-501. |
| [29] | Wedding RT. Malic enzymes of higher plants: characteristics, regulation, and physiological function [J]. Plant Physiol, 1989, 90(2): 367-371. |
| [30] | Bonnerot C, Galle AM, Jolliot A, et al. Purification and properties of plant cytochrome b5 [J]. Biochem J, 1985, 226(1): 331-334. |
| [31] | Kumar R, Tran LP, Neelakandan AK, et al. Higher plant cytochrome b5 polypeptides modulate fatty acid desaturation [J]. PLoS One, 2012, 7(2): e31370. |
| [32] | Schultz CJ, Coruzzi GM. The aspartate aminotransferase gene family of Arabidopsis encodes isoenzymes localized to three distinct subcellular compartments [J]. Plant J, 1995, 7(1): 61-75. |
| [33] | Allen DK, Young JD. Carbon and nitrogen provisions alter the metabolic flux in developing soybean embryos [J]. Plant Physiol, 2013, 161(3): 1458-1475. |
| [34] | Eastmond PJ, Dennis DT, Rawsthorne S. Evidence that a malate/inorganic phosphate exchange translocator imports carbon across the leucoplast envelope for fatty acid synthesis in developing castor seed endosperm [J]. Plant Physiol, 1997, 114(3): 851-856. |
| [35] | 李秀婷. 现代啤酒生产工艺 [M]. 北京: 中国农业大学出版社, 2013. |
| Li XT. Modern technology of beer production [M]. Beijing: China Agricultural University Press, 2013. | |
| [36] | 荆美玲. 油莎豆油脂合成基因CePDAT3基因的克隆及功能研究 [D]. 长春: 吉林农业大学, 2022. |
| Jing ML. Cloning and functional study of CePDAT3 gene for oil synthesis fromcyperus esculentusis [D]. Changchun: Jilin Agricultural University, 2022. | |
| [37] | Cramer CL, Weissenborn DL, Oishi KK, et al. Bioproduction of human enzymes in transgenic tobacco [J]. Ann N Y Acad Sci, 1996, 792(1): 62-71. |
| [38] | 吉夏洁. 过表达蓖麻RcWRI1提高烟叶油脂积累 [D]. 太谷: 山西农业大学, 2018. |
| Ji XJ. Overexpression of Ricinus communis RcWRI1 improves oil accumulation in tobacco leaves [D]. Taigu: Shanxi Agricultural University, 2018. | |
| [39] | Wilson RS, Thelen JJ. In vivo quantitative monitoring of subunit stoichiometry for metabolic complexes [J]. J Proteome Res, 2018, 17(5): 1773-1783. |
| [1] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [2] | YANG Wei-cheng, SUN Yan, YANG Qian, WANG Zhuang-lin, MA Ju-hua, XUE Jin-ai, LI Run-zhi. Genome-wide Identification of the FAX family in Gossypium hirsutum and Functional Analysis of GhFAX1 [J]. Biotechnology Bulletin, 2024, 40(3): 155-169. |
| [3] | LU Yu-dan, LIU Xiao-chi, FENG Xin, CHEN Gui-xin, CHEN Yi-ting. Identification of the Kiwifruit BBX Gene Family and Analysis of Their Transcriptional Characteristics [J]. Biotechnology Bulletin, 2024, 40(2): 172-182. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||