








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (10): 222-232.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0354
Previous Articles Next Articles
ZHANG Yu-qing( ), DONG Li-xue, ZHANG Bao-yue, ZHANG Ying, LIU Xue-ao, XIONG Shuang-xi(
), DONG Li-xue, ZHANG Bao-yue, ZHANG Ying, LIU Xue-ao, XIONG Shuang-xi( ), ZHANG Hong-xia(
), ZHANG Hong-xia( )
)
Received:2025-04-02
Online:2025-10-26
Published:2025-10-28
Contact:
XIONG Shuang-xi, ZHANG Hong-xia
E-mail:1924118220@qq.com;sxxiong@ldu.edu.cn;hxzhang@sibs.ac.cn
ZHANG Yu-qing, DONG Li-xue, ZHANG Bao-yue, ZHANG Ying, LIU Xue-ao, XIONG Shuang-xi, ZHANG Hong-xia. Characterization of the Activation Domain of SlMYB80 in Tomato and Its Function Validation during Pollen Development in Arabidopsis[J]. Biotechnology Bulletin, 2025, 41(10): 222-232.
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| β-tub qRT F | GATTTCAAAGATTAGGGAAGAGTA | RT-qPCR |
| β-tub qRT R | GTTCTGAAGCAAATGTCATAGAG | |
| SlMYB80 qRT F | ATTATCTGGTATGGGAATTGATCC | |
| SlMYB80 qRT R | TTAACATGACTTGTACTTGGTGCA | |
| SlMYB80fullCDS-BK F | catatggccatggaggccATGGGAAGAATTCCATGTTGTG | 酵母杂交 Yeast hybrid |
| SlMYB80fullCDS-BK R | aggtcgacggatccccggTCAAACCATTGGATTCATTAGAT | |
| SlMYB80fullCDS-AD F | atggccatggaggccagtATGGGAAGAATTCCATGTTGTG | |
| SlMYB80fullCDS-AD R | tcgatgcccacccgggtgTCAAACCATTGGATTCATTAGAT | |
| SlMYB80CDS(319 aa)-BK F | catatggccatggaggccATGGGAAGAATTCCATGTTGTG | |
| SlMYB80CDS(319 aa)-BK R | aggtcgacggatccccggTCAAGTATTGAACATAGTTGTTGATCC | |
| SlMYB80CDS(298 aa)-BK F | catatggccatggaggccATGGGAAGAATTCCATGTTGTG | |
| SlMYB80CDS(298 aa)-BK R | aggtcgacggatccccggTCAACAATTATCATTGCTAATTTTC | |
| SlMYB80CDS(272 aa)-BK F | catatggccatggaggccATGGGAAGAATTCCATGTTGTG | |
| SlMYB80CDS(272 aa)-BK R | aggtcgacggatccccggTCATGCTGTGCATGTACTTCCA | |
| SlMYB80CDS-35S-GFP F | gagctcggtacccggATGGGAAGAATTCCATGTTGTGA | 亚细胞定位 Subcellular localization |
| SlMYB80CDS-35S-GFP R | catgtcgactctagaAACCATTGGATTCATTAGATCATC | |
| ProAtMS188 F | ttcgagctcggtacccggAAGTTGTGTTTTTTCCCAAGTCA | 互补验证 Complementary verification |
| ProAtMS188 R | caccattctagaggatccTTCTTCTTTCTTTCTTTCTAGTTTTT | |
| SlMYB80 full CDS F | gaaagaaagaaagaagaaATGGGAAGAATTCCATGTTGTGA | |
| SlMYB80 full CDS R | cttgctcaccattctagaTCAAACCATTGGATTCATTAGATCATC | |
| pAtMS188-SlMYB80 JD F | ATCTGAATGTAAAAGGAGTTGACC | 转基因阳性植株鉴定 Transgenic positive plants identification |
| pAtMS188-SlMYB80 JD R | ATGTTGAGCAATGTAAGATGAAAG |
Table 1 Primers sequences used in this article
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| β-tub qRT F | GATTTCAAAGATTAGGGAAGAGTA | RT-qPCR |
| β-tub qRT R | GTTCTGAAGCAAATGTCATAGAG | |
| SlMYB80 qRT F | ATTATCTGGTATGGGAATTGATCC | |
| SlMYB80 qRT R | TTAACATGACTTGTACTTGGTGCA | |
| SlMYB80fullCDS-BK F | catatggccatggaggccATGGGAAGAATTCCATGTTGTG | 酵母杂交 Yeast hybrid |
| SlMYB80fullCDS-BK R | aggtcgacggatccccggTCAAACCATTGGATTCATTAGAT | |
| SlMYB80fullCDS-AD F | atggccatggaggccagtATGGGAAGAATTCCATGTTGTG | |
| SlMYB80fullCDS-AD R | tcgatgcccacccgggtgTCAAACCATTGGATTCATTAGAT | |
| SlMYB80CDS(319 aa)-BK F | catatggccatggaggccATGGGAAGAATTCCATGTTGTG | |
| SlMYB80CDS(319 aa)-BK R | aggtcgacggatccccggTCAAGTATTGAACATAGTTGTTGATCC | |
| SlMYB80CDS(298 aa)-BK F | catatggccatggaggccATGGGAAGAATTCCATGTTGTG | |
| SlMYB80CDS(298 aa)-BK R | aggtcgacggatccccggTCAACAATTATCATTGCTAATTTTC | |
| SlMYB80CDS(272 aa)-BK F | catatggccatggaggccATGGGAAGAATTCCATGTTGTG | |
| SlMYB80CDS(272 aa)-BK R | aggtcgacggatccccggTCATGCTGTGCATGTACTTCCA | |
| SlMYB80CDS-35S-GFP F | gagctcggtacccggATGGGAAGAATTCCATGTTGTGA | 亚细胞定位 Subcellular localization |
| SlMYB80CDS-35S-GFP R | catgtcgactctagaAACCATTGGATTCATTAGATCATC | |
| ProAtMS188 F | ttcgagctcggtacccggAAGTTGTGTTTTTTCCCAAGTCA | 互补验证 Complementary verification |
| ProAtMS188 R | caccattctagaggatccTTCTTCTTTCTTTCTTTCTAGTTTTT | |
| SlMYB80 full CDS F | gaaagaaagaaagaagaaATGGGAAGAATTCCATGTTGTGA | |
| SlMYB80 full CDS R | cttgctcaccattctagaTCAAACCATTGGATTCATTAGATCATC | |
| pAtMS188-SlMYB80 JD F | ATCTGAATGTAAAAGGAGTTGACC | 转基因阳性植株鉴定 Transgenic positive plants identification |
| pAtMS188-SlMYB80 JD R | ATGTTGAGCAATGTAAGATGAAAG |
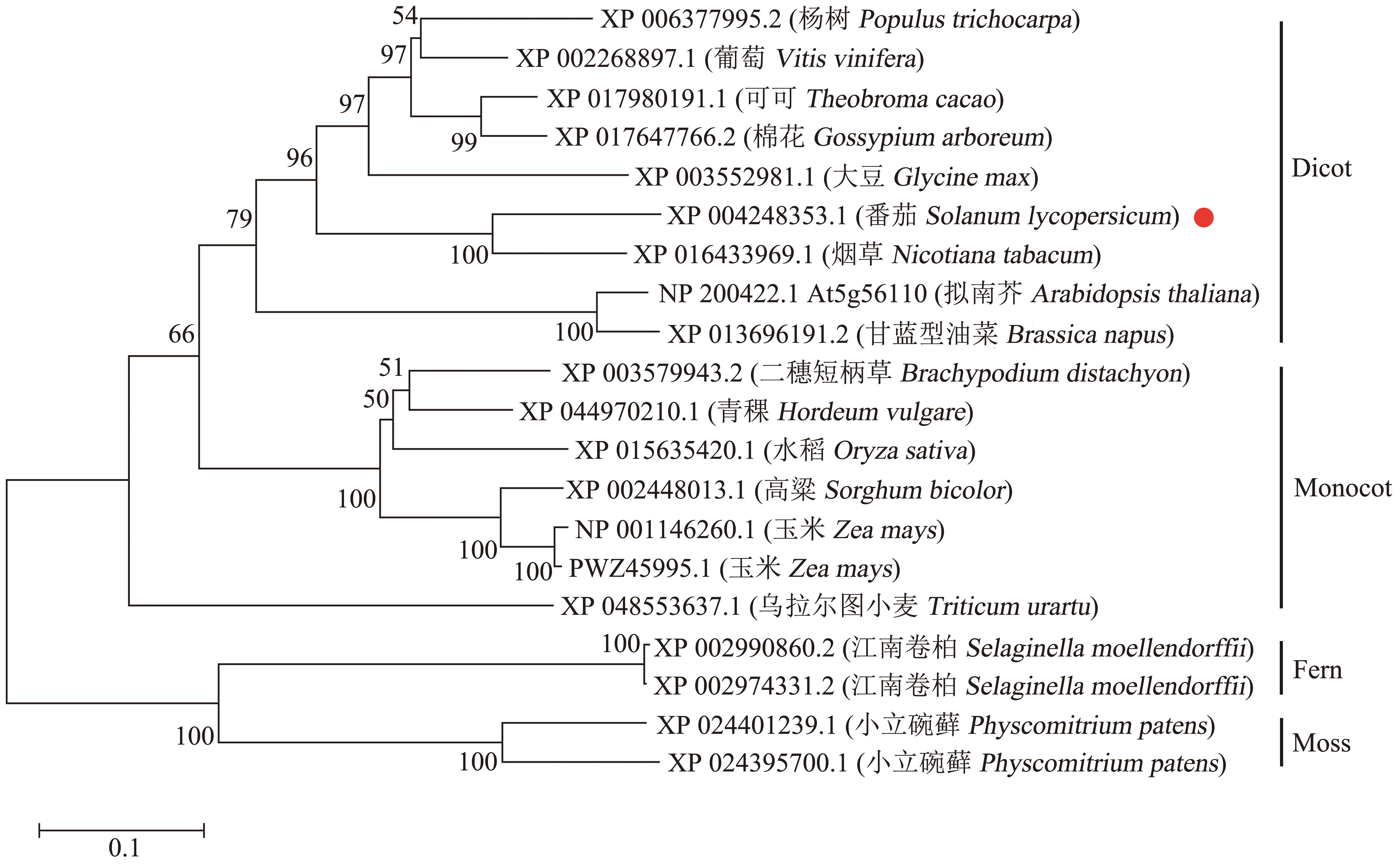
Fig. 1 Phylogenetic analysis of SlMYB80 with proteins from other speciesMonocot: Monocotyledon. Dicot: Dicotyledon. Red point indicates SlMYB80 from tomato

Fig. 2 PCR amplification of the full-length coding sequence (CDS) of SlMYB80 (A) and identification of positive clones by colony PCR assay (B)In Fig. A, 1: the full-length SlMYB80 CDS amplified from cDNA template; 2: negative control (ddH2O). In Fig. B, 1, 2: two single colonies of Escherichia coli; 3: positive control; 4: negative control (ddH2O). M: DL 5 000 marker

Fig. 3 SlMYB80-GFP protein specifically localized on the nucleus of tobacco mesophyll cellsA: Schematic diagram of subcellular localization construct. B‒E: Confocal microscopy images of p35s:GFP pCAMBIA1300 empty vector Agrobacterium-infiltrated tobacco leaves. F‒I: Confocal microscopy images of p35s:SlMYB80CDS-GFP pCAMBIA1300 Agrobacterium-infiltrated tobacco leaves. B and F: GFP signal channel. C and G: mCherry signal channel. D and H: Overlay signal channel images. E and I: Bright field channel images. Bar = 20 μm

Fig. 4 PCR amplification of diverse fragments of SlMYB80 CDS in tomato and colony PCR screening of positive clonesA: PCR amplification of region 1 (amino acids 1‒272) DNA fragment. B: PCR amplification of region 2 (amino acids 1‒298) DNA fragment. C: PCR amplification of region 3 (amino acids 1‒319) DNA fragment. D: PCR amplification of region 4 (amino acids 1‒336) DNA fragment. E‒H: positive clones detected by colony PCR of E. coli with different segments ligated into the pGBKT7 vector. In Fig. A‒D: M: DL 5 000 marker; 1: PCR amplification of different regions of the SlMYB80CDS DNA fragment; 2: negative control (ddH2O). In Fig. E‒H: M: DL 5 000 marker; 1‒2: two single colonies of E. coli; 3: positive control; 4: negative control (ddH2O)

Fig. 5 Activation domain of SlMYB80 on the 17 amino acid residues at the C-terminus of the polypeptide chainA: Schematic diagram of the connection of diverse fragments of SlMYB80 to the vectors pGBKT7 and empty pGADT7. B: Alignment of amino acid sequences between MS188/AtMYB80 and SlMYB80 (The red line indicates the R2R3 DNA-binding domain, and the red box indicates the activation domain). C: Identification of the activation of yeast reporter gene expression by diverse fragments of SlMYB80 on SD media lacking different amino acids. -Trp/Leu: SD medium without Trp and Leu; -Ade/-His/-Trp/-Leu: SD medium without Ade, His, Trp and Leu; -Ade/-His/-Trp/-Leu/X-α-gal: SD medium added X-α-gal without Ade, His, Trp and Leu

Fig. 6 Full-length coding sequence (CDS) of SlMYB80 can partially complement the phenotype of the Arabidopsis ms188A: Col-0 wild-type (WT). B: ms188 mutant. C: pAtMYB80:SlMYB80CDS p2300 complementation of the ms188; bar=2 cm; the white arrow indicates the recovered siliques. D‒F: Alexander staining (D: wild-type anther; E: ms188 anther; F: anther of the complemented plants (the white arrow indicates the recovered pollen grains); bar=100 μm). G: PCR identification of transgenic plants (DL2000 marker; 1‒5: independent transgenic lines; 6: positive control; 7: negative control (WT); 8: negative control (ddH2O)). H: RT-qPCR analysis of the relative expressions of SlMYB80 in transgenic plants. t-student test, *** indicates P<0.01
| [1] | 李君明, 项朝阳, 王孝宣, 等. “十三五” 我国番茄产业现状及展望 [J]. 中国蔬菜, 2021(2): 13-20. |
| Li JM, Xiang CY, Wang XX, et al. Current situation of tomato industry in China during ‘the thirteenth Five-Year Plan’ period and future prospect [J]. China Veg, 2021(2): 13-20. | |
| [2] | 胡雅丹, 伍国强, 刘晨, 等. MYB转录因子在调控植物响应逆境胁迫中的作用 [J]. 生物技术通报, 2024, 40(6): 5-22. |
| Hu YD, Wu GQ, Liu C, et al. Roles of MYB transcription factor in regulating the responses of plants to stress [J]. Biotechnol Bull, 2024, 40(6): 5-22. | |
| [3] | 李翔, 李涛, 宫超, 等. 番茄雄性不育研究现状与展望 [J]. 广东农业科学, 2023, 50(2): 49-58. |
| Li X, Li T, Gong C, et al. Current situation and prospect of research on male sterility of tomato [J]. Guangdong Agric Sci, 2023, 50(2): 49-58. | |
| [4] | Patikoglou G, Burley SK. Eukaryotic transcription factor-DNA complexes [J]. Annu Rev Biophys Biomol Struct, 1997, 26: 289-325. |
| [5] | Prouse MB, Campbell MM. The interaction between MYB proteins and their target DNA binding sites [J]. Biochim Biophys Acta Gene Regul Mech, 2012, 1819(1): 67-77. |
| [6] | Zhang ZB, Zhu J, Gao JF, et al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis [J]. Plant J, 2007, 52(3): 528-538. |
| [7] | Xiong SX, Lu JY, Lou Y, et al. The transcription factors MS188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana [J]. Plant J, 2016, 88(6): 936-946. |
| [8] | Wang K, Guo ZL, Zhou WT, et al. The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation [J]. Plant Physiol, 2018, 178(1): 283-294. |
| [9] | Han Y, Zhou SD, Fan JJ, et al. OsMS188 is a key regulator of tapetum development and sporopollenin synthesis in rice [J]. Rice, 2021, 14(1): 4. |
| [10] | He Y, Wang J, Hu JG, et al. Knockdown of SlEMS1 causes male sterility in tomato [J]. Hortic Plant J, 2025, 11(2): 939-942. |
| [11] | Jeong HJ, Kang JH, Zhao MA, et al. Tomato male sterile 1035 is essential for pollen development and meiosis in anthers [J]. J Exp Bot, 2014, 65(22): 6693-6709. |
| [12] | Sun ZL, Cheng BH, Zhang YH, et al. SlTDF1: a key regulator of tapetum degradation and pollen development in tomato [J]. Plant Sci, 2025, 351: 112321. |
| [13] | Bao HH, Ding YM, Yang F, et al. Gene silencing, knockout and over-expression of a transcription factor ABORTED MICROSPORES (SlAMS) strongly affects pollen viability in tomato (Solanum lycopersicum) [J]. BMC Genomics, 2022, 23(): 346. |
| [14] | Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11 [J]. Mol Biol Evol, 2021, 38(7): 3022-3027. |
| [15] | Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees [J]. Mol Biol Evol, 1987, 4(4): 406-425. |
| [16] | He SC, Ma R, Liu ZJ, et al. Overexpression of BnaAGL11, a MADS-box transcription factor, regulates leaf morphogenesis and senescence in Brassica napus [J]. J Agric Food Chem, 2022, 70(11): 3420-3434. |
| [17] | Li ZJ, Peng RH, Tian YS, et al. Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum [J]. Plant Cell Physiol, 2016, 57(8): 1657-1677. |
| [18] | 杨晶婷, 余艳, 高晓蓉, 等. MYB类转录因子调控植物耐逆机制的研究进展 [J]. 杭州师范大学学报: 自然科学版, 2021, 20(6): 621-627. |
| Yang JT, Yu Y, Gao XR, et al. Advances in plant MYB transcription factors regulation mechanisms to various stresses [J]. J Hangzhou Norm Univ Nat Sci Ed, 2021, 20(6): 621-627. | |
| [19] | Fang Q, Wang Q, Mao H, et al. AtDIV2, an R-R-type MYB transcription factor of Arabidopsis, negatively regulates salt stress by modulating ABA signaling [J]. Plant Cell Rep, 2018, 37(11): 1499-1511. |
| [20] | Zhang X, Chen LC, Shi QH, et al. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato [J]. Plant Sci, 2020, 291: 110356. |
| [21] | Wang ML, Hao J, Chen XH, et al. SlMYB102 expression enhances low-temperature stress resistance in tomato plants [J]. PeerJ, 2020, 8: e10059. |
| [22] | Liu XJ, Ban Z, Yang YY, et al. The yellowhorn MYB transcription factor MYB30 is required for wax accumulation and drought tolerance [J]. Tree Physiol, 2024, 44(10): tpae111. |
| [23] | Zhang Y, Zhang B, Yang TW, et al. The GAMYB-like gene SlMYB33 mediates flowering and pollen development in tomato [J]. Hortic Res, 2020, 7: 133. |
| [24] | Stevens VA, Murray BG. Studies on heteromorphic self-incompatibility systems: the cytochemistry and ultrastructure of the tapetum of Primula obconica [J]. J Cell Sci, 1981, 50: 419-431. |
| [25] | Vizcay-Barrena G, Wilson ZA. Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant [J]. J Exp Bot, 2006, 57(11): 2709-2717. |
| [26] | Parish RW, Li SF. Death of a tapetum: a programme of developmental altruism [J]. Plant Sci, 2010, 178(2): 73-89. |
| [27] | Fu ZZ, Yu J, Cheng XW, et al. The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development [J]. Plant Cell, 2014, 26(4): 1512-1524. |
| [28] | Wang N, Li X, Zhu J, et al. Molecular and cellular mechanisms of photoperiod- and thermo-sensitive genic male sterility in plants [J]. Mol Plant, 2025, 18(1): 26-41. |
| [29] | Gu JN, Zhu J, Yu Y, et al. DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis [J]. Plant J, 2014, 80(6): 1005-1013. |
| [30] | Lou Y, Xu XF, Zhu J, et al. The tapetal AHL family protein TEK determines nexine formation in the pollen wall [J]. Nat Commun, 2014, 5: 3855. |
| [31] | Phan HA, Iacuone S, Li SF, et al. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana [J]. Plant Cell, 2011, 23(6): 2209-2224. |
| [32] | Zhu J, Lou Y, Xu XF, et al. A genetic pathway for tapetum development and function in Arabidopsis [J]. J Integr Plant Biol, 2011, 53(11): 892-900. |
| [33] | Ghelli R, Brunetti P, Marzi D, et al. The full-length auxin response factor 8 isoform ARF8.1 controls pollen cell wall formation and directly regulates TDF1, AMS and MS188 expression [J]. Plant J, 2023, 113(4): 851-865. |
| [34] | Ko SS, Li MJ, Ho YC, et al. Rice transcription factor GAMYB modulates bHLH142 and is homeostatically regulated by TDR during anther tapetal and pollen development [J]. J Exp Bot, 2021, 72(13): 4888-4903. |
| [35] | Lu JY, Xiong SX, Yin WZ, et al. MS1 a direct target of MS188 regulates the expression of key sporophytic pollen coat protein genes in Arabidopsis [J]. J Exp Bot, 2020, 71(16): 4877-4889. |
| [36] | Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development [J]. Plant Cell, 2005, 17(3): 705-721. |
| [37] | 朱骏, 杨俊, 王晨, 等. 拟南芥转录因子AtMYB103的转录激活域鉴定 [J]. 上海交通大学学报: 农业科学版, 2009, 27(6): 611-614. |
| Zhu J, Yang J, Wang C, et al. Characterization of activation domain of the Arabidopsis transcription factor AtMYB103 [J]. J Shanghai Jiao Tong Univ Agric Sci, 2009, 27(6): 611-614. |
| [1] | CHEN Qiang, YU Ying-fei, ZHANG Ying, ZHANG Chong. Regulatory Effect of Methyl Jasmonate on Postharvest Chilling Injury in Oriental Melon ‘Emerald’ [J]. Biotechnology Bulletin, 2025, 41(9): 105-114. |
| [2] | SU Xiu-min, HAN Wen-qing, WANG Jiao, LI Peng, WANG Qiu-lan, LI Wan-xing, CAO Jin-jun. Isolation, Identification, Biological Characteristics and Biocontrol Effects of Trichoderma harzianum M408 against Tomato Early Blight [J]. Biotechnology Bulletin, 2025, 41(9): 277-288. |
| [3] | WANG Bin, LIN Chong, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui. Cloning of bHLH Transcription Factor UNE10 and Its Regulatory Roles in the Biosynthesis of Volatile Compounds in Clove Basil [J]. Biotechnology Bulletin, 2025, 41(9): 207-218. |
| [4] | HUANG Shi-yu, TIAN Shan-shan, YANG Tian-wei, GAO Man-rong, ZHANG Shang-wen. Genome-wide Identification and Expression Pattern Analysis of WRI1 Gene Family in Erythropalum scandens [J]. Biotechnology Bulletin, 2025, 41(8): 242-254. |
| [5] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [6] | LI Kai-jie, WU Yao, LI Dan-dan. Cloning of Gene CtbHLH128 in Safflower and Response Function Regulating Drought Stress [J]. Biotechnology Bulletin, 2025, 41(8): 234-241. |
| [7] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [8] | LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 181-192. |
| [9] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [10] | LI Rui, HU Ting, CHEN Shu-wei, WANG Yao, WANG Ji-ping. Positive Regulation of Anthocyanin Biosynthesis by PfMYB80 Transcription Factor in Perilla frutescens [J]. Biotechnology Bulletin, 2025, 41(6): 243-255. |
| [11] | GUO Tao, AI Li-jiao, ZOU Shi-hui, ZHOU Ling, LI Xue-mei. Functional Study of CjRAV1 from Camellia japonica in Regulating Flowering Delay [J]. Biotechnology Bulletin, 2025, 41(6): 208-217. |
| [12] | CHENG Shan, WANG Hui-wei, CHEN Chen, ZHU Ya-jing, LI Chun-xin, BIE Hai, WANG Shu-feng, CHEN Xian-gong, ZHANG Xiang-ge. Cloning of MYB Transcription Factor Gene CeMYB154 and Analysis of Salt Tolerance Function in Cyperus esculentus [J]. Biotechnology Bulletin, 2025, 41(6): 218-228. |
| [13] | CHENG Hui-juan, WANG Xin, SHI Xiao-tao, MA Dong-xu, GONG Da-chun, HU Jun-peng, XIE Zhi-wen. Effects of Transcription Factor CREA Knockout on the Morphology and the Secretion of β-glucosidase in Aspergillus niger [J]. Biotechnology Bulletin, 2025, 41(6): 344-354. |
| [14] | LI Xiao-huan, CHEN Xiang-yu, TAO Qi-yu, ZHU Ling, TANG Ming, YAO Yin-an, WANG Li-jun. Effects of PtoMYB61 on Lignin Biosynthesis and Salt Tolerance in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(6): 284-296. |
| [15] | LUO Si-fang, ZHANG Zu-ming, XIE Li-fang, GUO Zi-jing, CHEN Zhao-xing, YANG Yue-hua, YAN Xiang, ZHANG Hong-ming. Genome-wide Identification of GATA Gene Family of Jindou Kumquat (Fortunella hindsii) and Their Expression Analysis in Fruit Development [J]. Biotechnology Bulletin, 2025, 41(5): 218-230. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||