








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (7): 181-192.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0105
Previous Articles Next Articles
LI Xia( ), ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei(
), ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei( )
)
Received:2025-01-24
Online:2025-07-26
Published:2025-07-22
Contact:
JIAN Lei
E-mail:599570370@qq.com;jianleijane@imust.edu.cn
LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato[J]. Biotechnology Bulletin, 2025, 41(7): 181-192.
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 用途 Usage |
|---|---|---|
| StMYB96-F | CGACGACAAGACCGTGACCATGGGAAGACCACCTTGCTG | 基因克隆 |
| StMYB96-R | GAGGAGAAGAGCCGTCGAAAAAAGTCAGCAGATTCAC | Gene cloning |
| StMYB96-qRT-F | TCAAAGGGACAGTGGGAGAG | 荧光定量PCR |
| StMYB96-qRT-R | AGAGCTGGATCCTGTAACGG | RT-qPCR |
| StEF1α-qRT-F | GACAAGCGTGTTATTGAGAGG | 内参基因 |
| StEF1α-qRT-R | CACAGTGCAGTAGTACTTAGTG | Reference gene |
| StMYB96-OX-F | CACGGGGGACTCTAGAATGGGAAGACCACCTTGCTG | 过表达载体的构建 |
| StMYB96-OX-R | GGGGAAATTCGAGCTCTCAAAAAAAGTCAGCAGATT | Construction of overexpression vector |
Table 1 Primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 用途 Usage |
|---|---|---|
| StMYB96-F | CGACGACAAGACCGTGACCATGGGAAGACCACCTTGCTG | 基因克隆 |
| StMYB96-R | GAGGAGAAGAGCCGTCGAAAAAAGTCAGCAGATTCAC | Gene cloning |
| StMYB96-qRT-F | TCAAAGGGACAGTGGGAGAG | 荧光定量PCR |
| StMYB96-qRT-R | AGAGCTGGATCCTGTAACGG | RT-qPCR |
| StEF1α-qRT-F | GACAAGCGTGTTATTGAGAGG | 内参基因 |
| StEF1α-qRT-R | CACAGTGCAGTAGTACTTAGTG | Reference gene |
| StMYB96-OX-F | CACGGGGGACTCTAGAATGGGAAGACCACCTTGCTG | 过表达载体的构建 |
| StMYB96-OX-R | GGGGAAATTCGAGCTCTCAAAAAAAGTCAGCAGATT | Construction of overexpression vector |

Fig. 1 Cloning and sequence analysis of StMYB96 in S. tuberosum L.A: Electrophoretogram of PCR product of StMYB96 gene in S. tuberosum L. (M: DNA marker; P: StMYB96 PCR amplified product). B: Gene structure of StMYB96. C: Conserved domain analysis ofStMYB96 protein. D: StMYB96 gene sequence and deduced amino acid sequence (Green marked nucleotide sequence and pink marked amino acid sequence indicate MYB domain; the asterisk (*) refers to stop codon)

Fig. 2 Prediction of secondary and tertiary structure of StMYB96 proteinA: The secondary structure prediction of StMYB96 protein. Capital letters indicate amino acid sequence of StMYB96 protein. Lowercase letters indicate different secondary structures, where c, h, e and t indicate random coil, α-helix, extended strand and β-turn, respectively. B: The tertiary structure prediction of StMYB96 protein

Fig. 3 Multiple sequence alignment between StMYB96 and its homologous proteins from other speciesDark blue indicates amino acids completely conserved, pink indicates amino acids partially conserved, and light blue indicates amino acids similar

Fig. 4 Phylogenetic tree of StMYB96 proteinStMYB96: Solanum tuberosum (PGSC0003DMG400019535); SlMYB306: Solanum lycopersicum (XP_004236011.1); CaMYB306: Capsicum annuum (XP_016563223.1); LbMYB306b: Lycium barbarum (XP_060205081.1); NtMYB306: Nicotiana tabacum (XP_016452464.1); ArMYB96: Actinidia rufa (GFZ17560.1); AtMYB96: Arabidopsis thaliana (NP_201053.2); AtMYB94 (OAP04445.1)

Fig. 5 Subcellular localization of StMYB96 in S. tuberosum L.A: The structure of 35S::StMYB96::GFP vector. B: Subcellular localization of GFP protein and StMYB96::GFP fusion protein in Nicotiana benthamiana epidermal cells. (From left to right: GFP fluorescence, chloroplast self-luminescence, bright-field, and merged microscope image; Scale bar = 50 µm)

Fig. 6 Expression analysis of StMYB96 in various tissues and in response to various abiotic stressesA: Relative expressions of StMYB96 in different tissues. B: Expression profiles of StMYB96 in response to drought stress. C: Expression profiles of StMYB96 in response to low temperature stress. HT: Hours of treatment. ** indicates significant difference at P<0.01
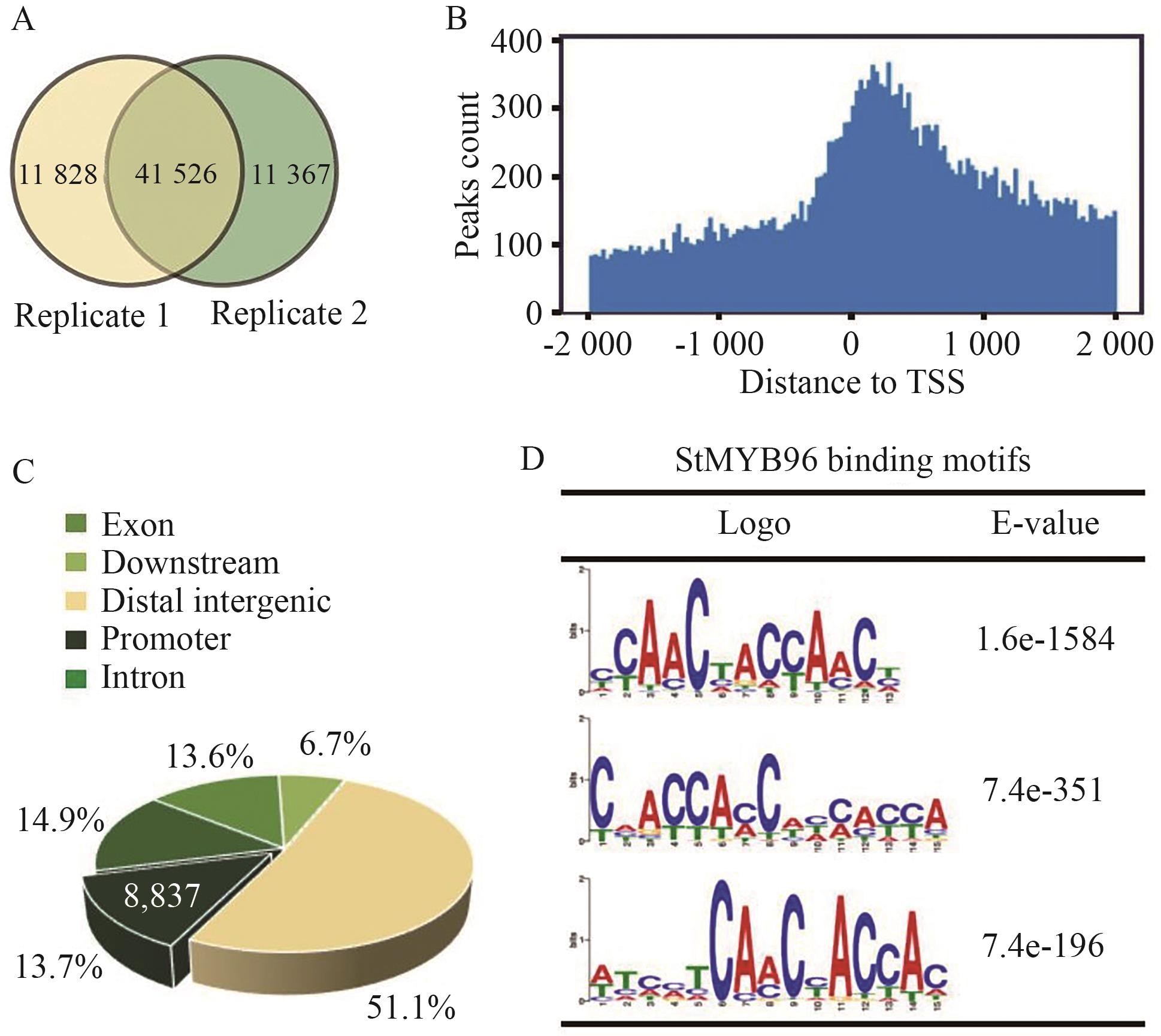
Fig. 7 Genome-wide identification of direct binding sites of StMYB96A: Venn diagram of peak number specifically binding to genomic DNA by StMYB96 identified in two biological replicates. B: Distribution of StMYB96 binding sites related to transcription start sites (TSS ±2 000 bp). C: Distribution of StMYB96 binding sites in potato genome. D: Binding motif of StMYB96

Fig. 8 KEGG pathway enrichment analysis and GO functional annotation analysis of StMYB96 target genesA: KEGG pathway enrichment analysis of StMYB96 target genes. The x-axis refers to the Rich factor corresponding to the pathways, the y-axis refers to the pathway names, the sizes of the dots indicate the number of target genes in each pathway, and the color of the dot indicates the size of the P-value. B: GO functional annotation analysis of StMYB96 target genes. The x-axis refers to the next-level GO terms under the three main GO categories (biological process, molecular function, and cellular component), and the y-axis refers to the proportion of target genes annotated to each term

Fig. 9 StMYB96-target genes involved in the responses to drought and low-temperature stressA: StMYB96-target genes involved in the flavonoid biosynthesis and fatty acid elongation pathway. B: StMYB96-target genes involved in the CBF signaling pathway. C: StMYB96-target genes involved in some functional annotations and drought stress response. Rep1 and Rep2 refer to two biological replicates. Fold enrichment indicates the binding enrichment level of StMYB96 in the promoter regions of genes

Fig. 10 A working model hypothesis of StMYB96 in response to drought and low-temperature stress in potatoSolid lines indicate known or experimentally validated regulatory pathways, dashed lines indicate hypothesized regulatory pathways, arrows indicate activation, and T-shaped arrows indicate inhibition
| [1] | Wang XP, Niu YL, Zheng Y. Multiple functions of MYB transcription factors in abiotic stress responses [J]. Int J Mol Sci, 2021, 22(11): 6125. |
| [2] | Wu XY, Xia M, Su P, et al. MYB transcription factors in plants: a comprehensive review of their discovery, structure, classification, functional diversity and regulatory mechanism [J]. Int J Biol Macromol, 2024, 282(Pt 2): 136652. |
| [3] | 位欣欣, 兰海燕. 植物MYB转录因子调控次生代谢及逆境响应的研究进展 [J]. 生物技术通报, 2022, 38(8): 12-23. |
| Wei XX, Lan HY. Advances in the regulation of plant MYB transcription factors in secondary metabolism and stress response [J]. Biotechnol Bull, 2022, 38(8): 12-23. | |
| [4] | 胡雅丹, 伍国强, 刘晨, 等. MYB转录因子在调控植物响应逆境胁迫中的作用 [J]. 生物技术通报, 2024, 40(6): 5-22. |
| Hu YD, Wu GQ, Liu C, et al. The role of MYB transcription factor in regulating plant response to adversity stress [J]. China Ind Econ, 2024, 40(6): 5-22. | |
| [5] | Yadav B, Jogawat A, Rahman MS, et al. Secondary metabolites in the drought stress tolerance of crop plants: a review [J]. Gene Rep, 2021, 23: 101040. |
| [6] | Sudiro C, Guglielmi F, Hochart M, et al. A phenomics and metabolomics investigation on the modulation of drought stress by a biostimulant plant extract in tomato (Solanum lycopersicum) [J]. Agronomy, 2022, 12(4): 764. |
| [7] | Qin LM, Sun L, Wei L, et al. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress [J]. Plant J, 2021, 105(4): 1010-1025. |
| [8] | Zhang L, Wang L, Fang YC, et al. Phosphorylated transcription factor PuHB40 mediates ROS-dependent anthocyanin biosynthesis in pear exposed to high light [J]. Plant Cell, 2024, 36(9): 3562-3583. |
| [9] | Naing AH, Kim CK. Abiotic stress-induced anthocyanins in plants: their role in tolerance to abiotic stresses [J]. Physiol Plant, 2021, 172(3): 1711-1723. |
| [10] | Maier A, Schrader A, Kokkelink L, et al. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis [J]. Plant J, 2013, 74(4): 638-651. |
| [11] | Kumar M, Kumar Patel M, Kumar N, et al. Metabolomics and molecular approaches reveal drought stress tolerance in plants [J]. Int J Mol Sci, 2021, 22(17): 9108. |
| [12] | Jiang H, Zhou LJ, Gao HN, et al. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple [J]. Plant Physiol, 2022, 189(4): 2044-2060. |
| [13] | Wang FB, Kong WL, Wong G, et al. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana [J]. Mol Genet Genomics, 2016, 291(4): 1545-1559. |
| [14] | Chen XS, Wu Y, Yu ZH, et al. BcMYB111 responds to BcCBF2 and induces flavonol biosynthesis to enhance tolerance under cold stress in non-heading Chinese cabbage [J]. Int J Mol Sci, 2023, 24(10): 8670. |
| [15] | Wang FB, Ren XQ, Zhang F, et al. A R2R3-type MYB transcription factor gene from soybean, GmMYB12, is involved in flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis [J]. Plant Biotechnol Rep, 2019, 13(3): 219-233. |
| [16] | Liu TL, Chen TZ, Kan JL, et al. The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants [J]. Plant Biotechnol J, 2022, 20(4): 722-735. |
| [17] | Chen YN, Feng PP, Zhang XW, et al. Silencing of SlMYB50 affects tolerance to drought and salt stress in tomato [J]. Plant Physiol Biochem, 2022, 193: 139-152. |
| [18] | Chen YN, Li L, Tang BY, et al. Silencing of SlMYB55 affects plant flowering and enhances tolerance to drought and salt stress in tomato [J]. Plant Sci, 2022, 316: 111166. |
| [19] | Lee SB, Kim HU, Suh MC. MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis [J]. Plant Cell Physiol, 2016, 57(11): 2300-2311. |
| [20] | Jian L, Kang K, Choi Y, et al. Mutation of OsMYB60 reduces rice resilience to drought stress by attenuating cuticular wax biosynthesis [J]. Plant J, 2022, 112(2): 339-351. |
| [21] | Song Q, Kong LF, Yang XR, et al. PtoMYB142, a poplar R2R3-MYB transcription factor, contributes to drought tolerance by regulating wax biosynthesis [J]. Tree Physiol, 2022, 42(10): 2133-2147. |
| [22] | Cao MX, Liu HZ, Zhang C, et al. Functional analysis of StPHT1;7, a Solanum tuberosum L. phosphate transporter gene, in growth and drought tolerance [J]. Plants, 2020, 9(10): 1384. |
| [23] | Ma X, Yu YN, Jia JH, et al. The pepper MYB transcription factor CaMYB306 accelerates fruit coloration and negatively regulates cold resistance [J]. Sci Hortic, 2022, 295: 110892. |
| [24] | Yu J, Lei B, Zhao HN, et al. Cloning, characterization and functional analysis of NtMYB306a gene reveals its role in wax alkane biosynthesis of tobacco trichomes and stress tolerance [J]. Front Plant Sci, 2022, 13: 1005811. |
| [25] | Yeats TH, Rose JKC. The formation and function of plant cuticles [J]. Plant Physiol, 2013, 163(1): 5-20. |
| [26] | Lian XY, Gao HN, Jiang H, et al. MdKCS2 increased plant drought resistance by regulating wax biosynthesis [J]. Plant Cell Rep, 2021, 40(12): 2357-2368. |
| [27] | Wu JT, Lv SD, Zhao L, et al. Advances in the study of the function and mechanism of the action of flavonoids in plants under environmental stresses [J]. Planta, 2023, 257(6): 108. |
| [28] | 林春草, 陈大伟, 戴均贵. 黄酮类化合物合成生物学研究进展 [J]. 药学学报, 2022, 57(5): 1322-1335. |
| Lin CC, Chen DW, Dai JG. Advances of synthetic biology of flavonoids [J]. Acta Pharm Sin, 2022, 57(5): 1322-1335. | |
| [29] | Liu T, Liu L, Zhou TS, et al. Chalcone isomerase gene (OsCHI3) increases rice drought tolerance by scavenging ROS via flavonoid and ABA metabolic pathways [J]. Crop J, 2025, 13(2): 372-384. |
| [30] | Bao HH, Yuan L, Luo YC, et al. The transcription factor WRKY41-FLAVONOID 3'-HYDROXYLASE module fine-tunes flavonoid metabolism and cold tolerance in potato [J]. Plant Physiol, 2025, 197(3): kiaf070. |
| [31] | Huang L, Hong YB, Zhang HJ, et al. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance [J]. BMC Plant Biol, 2016, 16(1): 203. |
| [32] | Chen YH, Hu L, Punta M, et al. Homologue structure of the SLAC1 anion channel for closing stomata in leaves [J]. Nature, 2010, 467(7319): 1074-1080. |
| [33] | Sun SJ, Qi GN, Gao QF, et al. Protein kinase OsSAPK8 functions as an essential activator of S-type anion channel OsSLAC1, which is nitrate-selective in rice [J]. Planta, 2016, 243(2): 489-500. |
| [34] | Riboni M, Galbiati M, Tonelli C, et al. GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS [J]. Plant Physiol, 2013, 162(3): 1706-1719. |
| [35] | Lim C, Kang K, Shim Y, et al. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways [J]. Plant Physiol, 2022, 188(4): 1900-1916. |
| [36] | Wang F, Chen HW, Li QT, et al. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants [J]. Plant J, 2015, 83(2): 224-236. |
| [37] | Luo X, Bai X, Sun XL, et al. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling [J]. J Exp Bot, 2013, 64(8): 2155-2169. |
| [1] | ZHANG Xue-qiong, PAN Su-jun, LI Wei, DAI Liang-ying. Research Progress of Plant Phosphate Transporters in the Response to Stress [J]. Biotechnology Bulletin, 2025, 41(7): 28-36. |
| [2] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [3] | WANG Fang, QIAO Shuai, SONG Wei, CUI Peng-juan, LIAO An-zhong, TAN Wen-fang, YANG Song-tao. Genome-wide Identification of the IbNRT2 Gene Family and Its Expression in Sweet Potato [J]. Biotechnology Bulletin, 2025, 41(7): 193-204. |
| [4] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [5] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [6] | LUO Ji-lin, LI Jin-ye, JIA Yu-xin. Identification and Functional Analysis of Gravity Response Regulatory Genes in Potato [J]. Biotechnology Bulletin, 2025, 41(6): 109-118. |
| [7] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [8] | DUAN Yong-hong, YANG Xin, YU Guan-qun, XIA Jun-jun, SONG Lu-shuai, BAI Xiao-dong, PENG Suo-tang. Genetic Diversity and Principal Component Analysis of 125 Potato Germplasm Resources [J]. Biotechnology Bulletin, 2025, 41(6): 130-143. |
| [9] | GUO Tao, AI Li-jiao, ZOU Shi-hui, ZHOU Ling, LI Xue-mei. Functional Study of CjRAV1 from Camellia japonica in Regulating Flowering Delay [J]. Biotechnology Bulletin, 2025, 41(6): 208-217. |
| [10] | WU Hao, DONG Wei-feng, HE Zi-tian, LI Yan-xiao, XIE Hui, SUN Ming-zhe, SHEN Yang, SUN Xiao-li. Genome-wide Identification and Expression Analysis of the Rice BXL Gene Family [J]. Biotechnology Bulletin, 2025, 41(6): 87-98. |
| [11] | HUANG Dan, PENG Bing-yang, ZHANG Pan-pan, JIAO Yue, LYU Jia-bin. Identification of HD-Zip Gene Family in Camellia oleifera and Analysis of Its Expression under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(6): 191-207. |
| [12] | PENG Shao-zhi, WANG Deng-ke, ZHANG Xiang, DAI Xiong-ze, XU Hao, ZOU Xue-xiao. Cloning, Expression Characteristics and Functional Verification of the Pepper CaFD1 Gene [J]. Biotechnology Bulletin, 2025, 41(5): 153-164. |
| [13] | SONG Hui-yang, SU Bao-jie, LI Jing-hao, MEI Chao, SONG Qian-na, CUI Fu-zhu, FENG Rui-yun. Cloning and Functional Analysis of the StAS2-15 Gene in Potato under Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 119-128. |
| [14] | LIU Yuan, ZHAO Ran, LU Zhen-fang, LI Rui-li. Research Progress in the Biological Metabolic Pathway and Functions of Plant Carotenoids [J]. Biotechnology Bulletin, 2025, 41(5): 23-31. |
| [15] | HU Ruo-qun, ZENG Jing-jing, LIANG Wan-feng, CAO Jia-yu, HUANG Xiao-wei, LIANG Xiao-ying, QIU Ming-yue, CHEN Ying. Integrated Transcriptome and Metabolome Analysis to Explore the Carotenoid Synthesis and Metabolism Mechanism in Anoectochilus roxburghii under Different Shading Conditions [J]. Biotechnology Bulletin, 2025, 41(5): 231-243. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||