








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (4): 227-242.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0918
SONG Jia-yi( ), SU Yun-li, ZHENG Xing-yan, XIA Wen-nian, YANG Dong-mei, HU Hui-zhen(
), SU Yun-li, ZHENG Xing-yan, XIA Wen-nian, YANG Dong-mei, HU Hui-zhen( )
)
Received:2024-09-21
Online:2025-04-26
Published:2025-04-25
Contact:
HU Hui-zhen
E-mail:siklib-91@swfu.edu.cn;Jenny_8729@163.com
SONG Jia-yi, SU Yun-li, ZHENG Xing-yan, XIA Wen-nian, YANG Dong-mei, HU Hui-zhen. Identification of the Snapdragon Expansin Gene Family and Screening of Its Genes Related to Resistance to Sclerotinia sclerotiorum[J]. Biotechnology Bulletin, 2025, 41(4): 227-242.
| Gene name | Forward primer sequence (5′‒3′) | Reverse primer sequence (5′‒3′) |
|---|---|---|
| AmUBI(RT-qPCR) | ATCCACCCTTCACCTTGTG | TTGTCAATGGTATCCGAGC |
| AmEXPA1(RT-qPCR) | ATGTGCCGATAATTTTGGTC | CATCACTGCCTCCATAGAAG |
| AmEXPA2(RT-qPCR) | GACTATGGCGGATGGGAAAG | TACTGGTGCCGTATCCTTCG |
| AmEXPA3(RT-qPCR) | ACCAACACCCTCTCTCGCTA | TATTGACGCCGTAGCCTTGG |
| AmEXPA4(RT-qPCR) | TTCTCATTCTTTGCCTACTGC | ATAACCACAAGCACCTCCCAT |
| AmEXPA5(RT-qPCR) | TCGCCAATGCCAATGCTTTC | AATGCAGTGCTCAATGCTGC |
| AmEXPA6(RT-qPCR) | GTGATGCTTCCGGCACAATG | GTCCACATGATGCCCCACTA |
| AmEXPA7(RT-qPCR) | TTCGTCGCTCTTCAGCTTGT | CGCAACTGTACCCGTTGTTG |
| AmEXPA8(RT-qPCR) | GGCTTCTTTGCTTCATTCAT | TAGAATGTAGCGTATGCCTG |
| AmEXPA9(RT-qPCR) | GTTGAAGCTAGGATTCCCGG | CTTCCGCCGTAGAATGTAGC |
| AmEXPA10(RT-qPCR) | CTCTTGGTGGGTTTCTTATCC | CCTTGGCTGTAAAGATTCCC |
| AmEXPA11(RT-qPCR) | TCATCAATCTTCGTCAACTCTCAC | CTTGAGGGAGGAGGAGGAGTC |
| AmEXPA12(RT-qPCR) | GGTATGGTACAAACACAGCAG | ACCACCCTCCGTCACTATTTG |
| AmEXPA13(RT-qPCR) | ATATTGATAGTTGTACCATTTTGTT | GATGAATACAAGTCTCCATATCC |
| AmEXPA14(RT-qPCR) | GGGTTTTTTCGCAATGGTTTC | GTAGAAGGTGGCATGAGCGTC |
| AmEXPA15(RT-qPCR) | GGCTGCAGTCATCTGCTACT | ATCCACAGGCACCCTCATTG |
| AmEXPA16(RT-qPCR) | CCATATTAGCCTGCATTGCC | TCGCTCCCACCGTAGAATGT |
| AmEXPA17(RT-qPCR) | GTGGGGCTTGTGGATATGGA | TGAGCAAGGTCGAAGTGGTG |
| AmEXPA18(RT-qPCR) | CAAGGTCGTGGTTGGCTGAA | CCCCAAAACCAGCTCCATAT |
| AmEXPA19(RT-qPCR) | CAAGGTCGTGGTTGGCTGAA | CCCCAAAACCAGCTCCATAT |
| AmEXPA20(RT-qPCR) | CAAGGTCGTGGTTGGCTGAA | GCCCCAAAACCAGCTCCATA |
| AmEXPA21(RT-qPCR) | TTCTGCTTACGCGGTGCTTA | TAGGCACCATTTGGGGTCAC |
| AmEXPA22(RT-qPCR) | GTGGTGCTTGTGGGTATGG | AATCTCAAAGCATTCCCCAC |
| AmEXPA23(RT-qPCR) | CCTATCAGACGACGAAGGGC | CGTATCCACATGCTCCTCCC |
| AmEXPA24(RT-qPCR) | TGAAGCCACTAATCTGCATTCTTAG | GCCATAGAACGTAGCGTGTGC |
| AmEXPA25(RT-qPCR) | GGCCACTTCTCAGCTATTTGTCA | GGATCCACCATAAAATGTTGCAT |
| AmEXPB1(RT-qPCR) | TTGTTTCGTTTCCACTTCACTTTTT | AGTAACCAGAGAAGAAAATGGAGCC |
| Am EXPB2(RT-qPCR) | TGCTTTTGCATACCATCAGTG | AGAGGTAAAACTTCGTCTGGA |
| Am EXPB3(RT-qPCR) | CCCTCCTTCTCCGGTATTTC | AACGGCTTCACGTCCACCA |
| Am EXPB4(RT-qPCR) | TATAGCTACCTTGGTTGTTCAAATT | GAGCCCTAGTTACATCTTCTCTATA |
| AmEXLA1(RT-qPCR) | GGGTATGACGGGAAATGGTATT | AACCATCTTGAGCAATATCGGTAA |
| AmEXLB1(RT-qPCR) | TCAGCAAAATTGATGATTATTCTGC | CAATCGGGACTTCCATAATAGGTT |
| AmEXLB2(RT-qPCR) | ATGACTCGTGTTAGACTTTCTTTGA | TCCCAGATTACACATAAAAAGAAAA |
| AmEXPA2(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXPA12(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXPA9(CDS) | GACTTGAACGGT CACAAAGTTCCTTGAAAATG | TCGGGGAAATTC ATTTTTGCCCATCAGACG |
| AmEXPA13(CDS) | GACTTGAACGGT ATCTTTCTTCATTAGACCCT | TCGGGGAAATTC CGAAATCGAAAACAATAGCT |
| AmEXPA16(CDS) | GACTTGAACGGT TTTAAACCCCTCTCCATCTC | TCGGGGAAATTC TTGTATTAAAACTGAGCCCCT |
| AmEXPA17(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXPA21(CDS) | GACTTGAACGGT CTCTGTGTACCATTTTCTCC | TCGGGGAAATTC AATTCGACCCTATAACAAAGT |
| AmEXPB1(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXLB1(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXLB2(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
Table 1 Primers for RT-qPCR
| Gene name | Forward primer sequence (5′‒3′) | Reverse primer sequence (5′‒3′) |
|---|---|---|
| AmUBI(RT-qPCR) | ATCCACCCTTCACCTTGTG | TTGTCAATGGTATCCGAGC |
| AmEXPA1(RT-qPCR) | ATGTGCCGATAATTTTGGTC | CATCACTGCCTCCATAGAAG |
| AmEXPA2(RT-qPCR) | GACTATGGCGGATGGGAAAG | TACTGGTGCCGTATCCTTCG |
| AmEXPA3(RT-qPCR) | ACCAACACCCTCTCTCGCTA | TATTGACGCCGTAGCCTTGG |
| AmEXPA4(RT-qPCR) | TTCTCATTCTTTGCCTACTGC | ATAACCACAAGCACCTCCCAT |
| AmEXPA5(RT-qPCR) | TCGCCAATGCCAATGCTTTC | AATGCAGTGCTCAATGCTGC |
| AmEXPA6(RT-qPCR) | GTGATGCTTCCGGCACAATG | GTCCACATGATGCCCCACTA |
| AmEXPA7(RT-qPCR) | TTCGTCGCTCTTCAGCTTGT | CGCAACTGTACCCGTTGTTG |
| AmEXPA8(RT-qPCR) | GGCTTCTTTGCTTCATTCAT | TAGAATGTAGCGTATGCCTG |
| AmEXPA9(RT-qPCR) | GTTGAAGCTAGGATTCCCGG | CTTCCGCCGTAGAATGTAGC |
| AmEXPA10(RT-qPCR) | CTCTTGGTGGGTTTCTTATCC | CCTTGGCTGTAAAGATTCCC |
| AmEXPA11(RT-qPCR) | TCATCAATCTTCGTCAACTCTCAC | CTTGAGGGAGGAGGAGGAGTC |
| AmEXPA12(RT-qPCR) | GGTATGGTACAAACACAGCAG | ACCACCCTCCGTCACTATTTG |
| AmEXPA13(RT-qPCR) | ATATTGATAGTTGTACCATTTTGTT | GATGAATACAAGTCTCCATATCC |
| AmEXPA14(RT-qPCR) | GGGTTTTTTCGCAATGGTTTC | GTAGAAGGTGGCATGAGCGTC |
| AmEXPA15(RT-qPCR) | GGCTGCAGTCATCTGCTACT | ATCCACAGGCACCCTCATTG |
| AmEXPA16(RT-qPCR) | CCATATTAGCCTGCATTGCC | TCGCTCCCACCGTAGAATGT |
| AmEXPA17(RT-qPCR) | GTGGGGCTTGTGGATATGGA | TGAGCAAGGTCGAAGTGGTG |
| AmEXPA18(RT-qPCR) | CAAGGTCGTGGTTGGCTGAA | CCCCAAAACCAGCTCCATAT |
| AmEXPA19(RT-qPCR) | CAAGGTCGTGGTTGGCTGAA | CCCCAAAACCAGCTCCATAT |
| AmEXPA20(RT-qPCR) | CAAGGTCGTGGTTGGCTGAA | GCCCCAAAACCAGCTCCATA |
| AmEXPA21(RT-qPCR) | TTCTGCTTACGCGGTGCTTA | TAGGCACCATTTGGGGTCAC |
| AmEXPA22(RT-qPCR) | GTGGTGCTTGTGGGTATGG | AATCTCAAAGCATTCCCCAC |
| AmEXPA23(RT-qPCR) | CCTATCAGACGACGAAGGGC | CGTATCCACATGCTCCTCCC |
| AmEXPA24(RT-qPCR) | TGAAGCCACTAATCTGCATTCTTAG | GCCATAGAACGTAGCGTGTGC |
| AmEXPA25(RT-qPCR) | GGCCACTTCTCAGCTATTTGTCA | GGATCCACCATAAAATGTTGCAT |
| AmEXPB1(RT-qPCR) | TTGTTTCGTTTCCACTTCACTTTTT | AGTAACCAGAGAAGAAAATGGAGCC |
| Am EXPB2(RT-qPCR) | TGCTTTTGCATACCATCAGTG | AGAGGTAAAACTTCGTCTGGA |
| Am EXPB3(RT-qPCR) | CCCTCCTTCTCCGGTATTTC | AACGGCTTCACGTCCACCA |
| Am EXPB4(RT-qPCR) | TATAGCTACCTTGGTTGTTCAAATT | GAGCCCTAGTTACATCTTCTCTATA |
| AmEXLA1(RT-qPCR) | GGGTATGACGGGAAATGGTATT | AACCATCTTGAGCAATATCGGTAA |
| AmEXLB1(RT-qPCR) | TCAGCAAAATTGATGATTATTCTGC | CAATCGGGACTTCCATAATAGGTT |
| AmEXLB2(RT-qPCR) | ATGACTCGTGTTAGACTTTCTTTGA | TCCCAGATTACACATAAAAAGAAAA |
| AmEXPA2(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXPA12(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXPA9(CDS) | GACTTGAACGGT CACAAAGTTCCTTGAAAATG | TCGGGGAAATTC ATTTTTGCCCATCAGACG |
| AmEXPA13(CDS) | GACTTGAACGGT ATCTTTCTTCATTAGACCCT | TCGGGGAAATTC CGAAATCGAAAACAATAGCT |
| AmEXPA16(CDS) | GACTTGAACGGT TTTAAACCCCTCTCCATCTC | TCGGGGAAATTC TTGTATTAAAACTGAGCCCCT |
| AmEXPA17(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXPA21(CDS) | GACTTGAACGGT CTCTGTGTACCATTTTCTCC | TCGGGGAAATTC AATTCGACCCTATAACAAAGT |
| AmEXPB1(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXLB1(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
| AmEXLB2(CDS) | GACTTGAACGGT | TCGGGGAAATTC |
试剂 Reagent | 母液浓度 Concentration of mother liquor/(mol·L-1) | 工作浓度 Concentration of working solution/(mmol·L-1) |
|---|---|---|
| MgCl2 | 1 | 10 |
| 2-(N-吗啉代)乙磺酸(pH=5.7) | 1 | 10 |
| 乙酰丁香酮 | 0.2 | 0.2 |
Table 2 Ingredient of infiltration buffer
试剂 Reagent | 母液浓度 Concentration of mother liquor/(mol·L-1) | 工作浓度 Concentration of working solution/(mmol·L-1) |
|---|---|---|
| MgCl2 | 1 | 10 |
| 2-(N-吗啉代)乙磺酸(pH=5.7) | 1 | 10 |
| 乙酰丁香酮 | 0.2 | 0.2 |
基因名称 Gene name | 基因号 Gene ID | 最佳区比例 Most favoured region/% | 次允许区比例 Additional allowed region/% | 一般允许区比例 Generously allowed region/% | 不允许区比例 Disallowed region/% | 生成因子值 G-factor |
|---|---|---|---|---|---|---|
| AmEXPA1 | Am01g01950.T01 | 90.6 | 9.0 | 0.0 | 0.5 | -0.09 |
| AmEXPA2 | Am01g11840.T01 | 92.7 | 6.8 | 0.5 | 0.0 | -0.12 |
| AmEXPA3 | Am01g25390.T01 | 91.6 | 8.4 | 0.0 | 0.0 | -0.12 |
| AmEXPA4 | Am01g26640.T01 | 91.5 | 8.0 | 0.5 | 0.0 | -0.14 |
| AmEXPA5 | Am01g44550.T01 | 88.3 | 11.2 | 0.5 | 0.0 | -0.10 |
| AmEXPA6 | Am01g44560.T01 | 88.2 | 11.4 | 0.5 | 0.0 | -0.09 |
| AmEXPA7 | Am02g27260.T01 | 87.0 | 10.3 | 1.8 | 0.9 | -0.18 |
| AmEXPA8 | Am02g33780.T01 | 87.2 | 11.5 | 0.4 | 0.9 | -0.17 |
| AmEXPA9 | Am03g06850.T01 | 89.7 | 10.3 | 0.0 | 0.0 | -0.14 |
| AmEXPA10 | Am03g32100.T01 | 92.1 | 7.9 | 0.0 | 0.0 | -0.12 |
| AmEXPA11 | Am03g40550.T01 | 90.5 | 7.8 | 1.3 | 0.4 | -0.13 |
| AmEXPA12 | Am04g05250.T01 | 92.2 | 7.2 | 0.0 | 0.6 | -0.08 |
| AmEXPA13 | Am05g03930.T01 | 88.4 | 10.7 | 0.9 | 0.0 | -0.17 |
| AmEXPA14 | Am05g12940.T01 | 91.6 | 8.4 | 0.0 | 0.0 | -0.11 |
| AmEXPA15 | Am05g41720.T01 | 88.9 | 9.2 | 0.5 | 1.4 | -0.18 |
| AmEXPA16 | Am06g27420.T01 | 87.6 | 12.4 | 0.0 | 0.0 | -0.10 |
| AmEXPA17 | Am06g34470.T01 | 87.6 | 12.4 | 0.0 | 0.0 | -0.13 |
| AmEXPA18 | Am07g13940.T01 | 91.4 | 8.6 | 0.0 | 0.0 | -0.11 |
| AmEXPA19 | Am07g15820.T01 | 91.4 | 8.6 | 0.0 | 0.0 | 0.00 |
| AmEXPA20 | Am07g18170.T01 | 91.5 | 8.5 | 0.0 | 0.0 | -0.09 |
| AmEXPA21 | Am07g21430.T01 | 92.1 | 7.9 | 0.0 | 0.0 | -0.08 |
| AmEXPA22 | Am07g35680.T01 | 92.5 | 6.9 | 0.6 | 0.0 | -0.12 |
| AmEXPA23 | Am08g26500.T01 | 92.4 | 6.5 | 1.1 | 0.0 | -0.14 |
| AmEXPA24 | Am08g34570.T01 | 91.8 | 7.8 | 0.0 | 0.5 | -0.11 |
| AmEXPA25 | Am08g37200.T01 | 92.6 | 7.4 | 0.0 | 0.0 | -0.08 |
| AmEXPB1 | Am01g01300.T01 | 90.4 | 8.7 | 0.9 | 0.0 | -0.19 |
| AmEXPB2 | Am04g35520.T01 | 76.7 | 17.5 | 4.8 | 1.1 | -0.38 |
| AmEXPB3 | Am07g07660.T01 | 87.8 | 11.7 | 0.0 | 0.5 | -0.13 |
| AmEXPB4 | Am07g15580.T01 | 74.5 | 19.0 | 4.3 | 2.2 | -0.48 |
| AmEXLA1 | Am08g01880.T02 | 90.7 | 8.9 | 0.0 | 0.4 | -0.12 |
| AmEXLB1 | Am03g20060.T01 | 89.2 | 9.4 | 0.5 | 0.9 | -0.17 |
| AmEXLB2 | Am04g31120.T01 | 88.1 | 11.4 | 0.5 | 0.0 | -0.10 |
Table 3 3D structures prediction of AmEXPs protein
基因名称 Gene name | 基因号 Gene ID | 最佳区比例 Most favoured region/% | 次允许区比例 Additional allowed region/% | 一般允许区比例 Generously allowed region/% | 不允许区比例 Disallowed region/% | 生成因子值 G-factor |
|---|---|---|---|---|---|---|
| AmEXPA1 | Am01g01950.T01 | 90.6 | 9.0 | 0.0 | 0.5 | -0.09 |
| AmEXPA2 | Am01g11840.T01 | 92.7 | 6.8 | 0.5 | 0.0 | -0.12 |
| AmEXPA3 | Am01g25390.T01 | 91.6 | 8.4 | 0.0 | 0.0 | -0.12 |
| AmEXPA4 | Am01g26640.T01 | 91.5 | 8.0 | 0.5 | 0.0 | -0.14 |
| AmEXPA5 | Am01g44550.T01 | 88.3 | 11.2 | 0.5 | 0.0 | -0.10 |
| AmEXPA6 | Am01g44560.T01 | 88.2 | 11.4 | 0.5 | 0.0 | -0.09 |
| AmEXPA7 | Am02g27260.T01 | 87.0 | 10.3 | 1.8 | 0.9 | -0.18 |
| AmEXPA8 | Am02g33780.T01 | 87.2 | 11.5 | 0.4 | 0.9 | -0.17 |
| AmEXPA9 | Am03g06850.T01 | 89.7 | 10.3 | 0.0 | 0.0 | -0.14 |
| AmEXPA10 | Am03g32100.T01 | 92.1 | 7.9 | 0.0 | 0.0 | -0.12 |
| AmEXPA11 | Am03g40550.T01 | 90.5 | 7.8 | 1.3 | 0.4 | -0.13 |
| AmEXPA12 | Am04g05250.T01 | 92.2 | 7.2 | 0.0 | 0.6 | -0.08 |
| AmEXPA13 | Am05g03930.T01 | 88.4 | 10.7 | 0.9 | 0.0 | -0.17 |
| AmEXPA14 | Am05g12940.T01 | 91.6 | 8.4 | 0.0 | 0.0 | -0.11 |
| AmEXPA15 | Am05g41720.T01 | 88.9 | 9.2 | 0.5 | 1.4 | -0.18 |
| AmEXPA16 | Am06g27420.T01 | 87.6 | 12.4 | 0.0 | 0.0 | -0.10 |
| AmEXPA17 | Am06g34470.T01 | 87.6 | 12.4 | 0.0 | 0.0 | -0.13 |
| AmEXPA18 | Am07g13940.T01 | 91.4 | 8.6 | 0.0 | 0.0 | -0.11 |
| AmEXPA19 | Am07g15820.T01 | 91.4 | 8.6 | 0.0 | 0.0 | 0.00 |
| AmEXPA20 | Am07g18170.T01 | 91.5 | 8.5 | 0.0 | 0.0 | -0.09 |
| AmEXPA21 | Am07g21430.T01 | 92.1 | 7.9 | 0.0 | 0.0 | -0.08 |
| AmEXPA22 | Am07g35680.T01 | 92.5 | 6.9 | 0.6 | 0.0 | -0.12 |
| AmEXPA23 | Am08g26500.T01 | 92.4 | 6.5 | 1.1 | 0.0 | -0.14 |
| AmEXPA24 | Am08g34570.T01 | 91.8 | 7.8 | 0.0 | 0.5 | -0.11 |
| AmEXPA25 | Am08g37200.T01 | 92.6 | 7.4 | 0.0 | 0.0 | -0.08 |
| AmEXPB1 | Am01g01300.T01 | 90.4 | 8.7 | 0.9 | 0.0 | -0.19 |
| AmEXPB2 | Am04g35520.T01 | 76.7 | 17.5 | 4.8 | 1.1 | -0.38 |
| AmEXPB3 | Am07g07660.T01 | 87.8 | 11.7 | 0.0 | 0.5 | -0.13 |
| AmEXPB4 | Am07g15580.T01 | 74.5 | 19.0 | 4.3 | 2.2 | -0.48 |
| AmEXLA1 | Am08g01880.T02 | 90.7 | 8.9 | 0.0 | 0.4 | -0.12 |
| AmEXLB1 | Am03g20060.T01 | 89.2 | 9.4 | 0.5 | 0.9 | -0.17 |
| AmEXLB2 | Am04g31120.T01 | 88.1 | 11.4 | 0.5 | 0.0 | -0.10 |
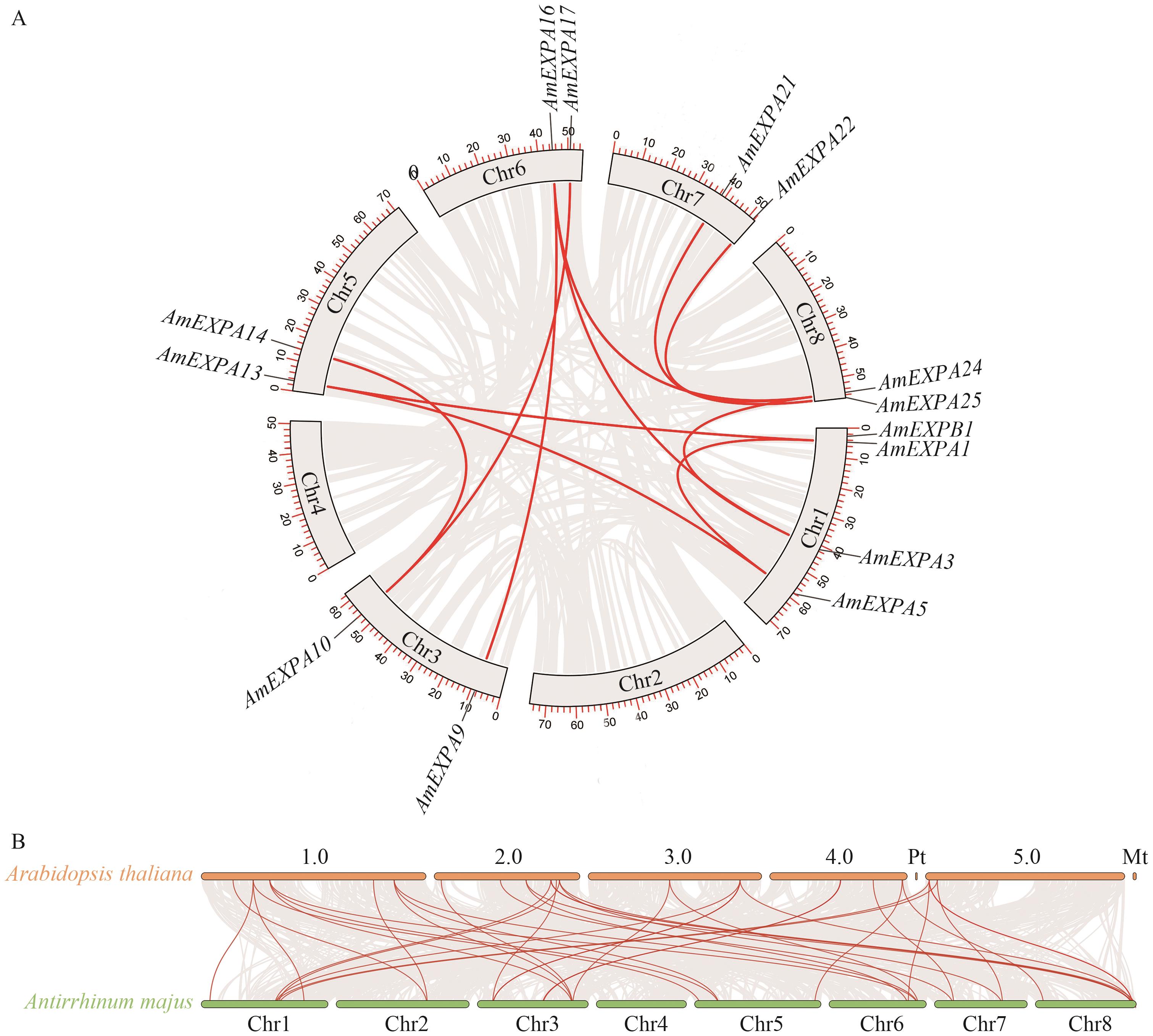
Fig. 5 Gene duplication (A) and interspecies collinearity analysis (B) of the AmEXPs familychr: Chromosome. The red line indicates the genomic and interspecific collinearity gene pairs of AmEXPs

Fig. 7 Phenotype observation and the areas of the lesions on the leaves of ‘Am1’ and ‘Am6’ in vitro inoculation of S. sclerotiorum and differentially expressed AmEXPs genes in RNA-seq analysisA: The phenotypes of ‘Am1’ (S) and ‘Am6’ (R) were observed after 0, 12, 18, 24, 36, and 42 h of infection with S. sclerotiorum, bars=1 cm. B: The areas of the lesions on the leaves in Fig. A. C: Differential expressed AmEXPs genes after 24 h inoculation with S. sclerotiorum. * P<0.05, ** P<0.01. hpi: Hours after inoculation. The same below

Fig. 10 Preliminary validation of AmEXPs candidate gene resistance in transient transformation of N. benthamianaA-J: Phenotype of Nicotiana benthamiana after transient transformation and Sclerotinia sclerotiorum infection for 36 h, bars=2 cm. K‒T: Statistical analysis of lesion area. U: Phylogenetic tree of AmEXPs protein family
| 1 | 陈宇华, 陈剑锋, 钟声远, 等. 20份金鱼草种质资源花色性状鉴定与分析 [J]. 福建农业科技, 2022, 53(7): 1-7. |
| Chen YH, Chen JF, Zhong SY, et al. Identification and analysis of flower color traits of 20 germplasm resources of Antirrhinum majus [J]. Fujian Agric Sci Technol, 2022, 53(7): 1-7. | |
| 2 | 齐伟. 金鱼草常见病虫害及防治 [J]. 花木盆景: 花卉园艺, 2002(9): 26. |
| Qi W. Common pests and diseases of snapdragon and their control [J]. Flowers Trees Potted Landsc, 2002(9): 26. | |
| 3 | 师莹莹, 李大勇, 张慧娟, 等. 植物细胞壁介导的抗病性及其分子机制 [J]. 植物生理学报, 2011, 47(7): 661-668. |
| Shi YY, Li DY, Zhang HJ, et al. Cell wall-mediated disease resistance and its molecular mechanism in plants [J]. Plant Physiol J, 2011, 47(7): 661-668. | |
| 4 | Cosgrove DJ. Plant expansins: diversity and interactions with plant cell walls [J]. Curr Opin Plant Biol, 2015, 25: 162-172. |
| 5 | Marowa P, Ding AM, Kong YZ. Expansins: roles in plant growth and potential applications in crop improvement [J]. Plant Cell Rep, 2016, 35(5): 949-965. |
| 6 | 赵美荣, 李永春, 王玮. 扩展蛋白与植物抗逆性关系研究进展 [J]. 植物生理学报, 2012, 48(7): 637-642. |
| Zhao MR, Li YC, Wang W. Research progress on relationship between expansin and plant resistance [J]. Plant Physiol J, 2012, 48(7): 637-642. | |
| 7 | Fu J, Liu HB, Li Y, et al. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice [J]. Plant Physiol, 2011, 155(1): 589-602. |
| 8 | Tan J, Wang ML, Shi ZY, et al. OsEXPA10 mediates the balance between growth and resistance to biotic stress in rice [J]. Plant Cell Rep, 2018, 37(7): 993-1002. |
| 9 | Abuqamar S, Ajeb S, Sham A, et al. A mutation in the expansin-like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana [J]. Mol Plant Pathol, 2013, 14(8): 813-827. |
| 10 | Nardi CF, Villarreal NM, Rossi FR, et al. Overexpression of the carbohydrate binding module of strawberry expansin2 in Arabidopsis thaliana modifies plant growth and cell wall metabolism [J]. Plant Mol Biol, 2015, 88(1/2): 101-117. |
| 11 | Chen LJ, Zou WS, Wu G, et al. Tobacco alpha-expansin EXPA4 plays a role in Nicotiana benthamiana defence against tobacco mosaic virus [J]. Planta, 2018, 247(2): 355-368. |
| 12 | 彭爱红, 张婧芸, 陈志毅, 等. CsEXPA8过表达对‘晚锦橙’生长及溃疡病抗性的影响 [J]. 园艺学报, 2024, 51(5): 971-981. |
| Peng AH, Zhang JY, Chen ZY, et al. Effects of overexpression of CsEXPA8 on growth and canker disease resistance in 'wanjincheng' orange [J]. Acta Hortic Sin, 2024, 51(5): 971-981. | |
| 13 | Brasileiro ACM, Lacorte C, Pereira BM, et al. Ectopic expression of an expansin-like B gene from wild Arachis enhances tolerance to both abiotic and biotic stresses [J]. Plant J, 2021, 107(6): 1681-1696. |
| 14 | 夏文念, 杨冬梅, 宋佳怡, 等. 金鱼草GAE基因家族鉴定及核盘菌抗性基因挖掘 [J]. 农业生物技术学报, 2024, 32(9): 2049-2059. |
| Xia WN, Yang DM, Song JY, et al. Identification of GAE gene family in Antirrhinum majus and mining of resistance genes to Sclerotinia sclerotiorum [J]. J Agric Biotechnol, 2024, 32(9): 2049-2059. | |
| 15 | 赵晗茜, 宋佳怡, 杨洁, 等. 金鱼草XTH家族基因鉴定及抗核盘菌和雄蕊瓣化相关基因筛选 [J]. 植物学报, 2024, 59(2): 188-203. |
| Zhao I, Song JY, Yang J, et al. Identification of XTH family genes in Antirrhinum majus and screening of genes involoved in Sclerotinia sclerotiorum resistance and stamen petalization [J]. Chin Bull Bot, 2024, 59(2): 188-203. | |
| 16 | Marchler-Bauer A, Bo Y, Han LY, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures [J]. Nucleic Acids Res, 2017, 45(D1): D200-D203. |
| 17 | Sánchez MA, Mateos I, Labrador E, et al. Brassinolides and IAA induce the transcription of four α-expansin genes related to development in Cicer arietinum [J]. Plant Physiol Biochem, 2004, 42(9): 709-716. |
| 18 | Chen YH, Xie B, An XH, et al. Overexpression of the apple expansin-like gene MdEXLB1 accelerates the softening of fruit texture in tomato [J]. J Integr Agric, 2022, 21(12): 3578-3588. |
| 19 | Muthusamy M, Kim JY, Yoon EK, et al. BrEXLB1, a Brassica rapa expansin-like B1 gene is associated with root development, drought stress response, and seed germination [J]. Genes, 2020, 11(4): 404. |
| 20 | Otulak-Kozieł K, Kozieł E, Lockhart BEL, et al. The expression of potato expansin A3 (StEXPA3) and Extensin4 (StEXT4) genes with distribution of StEXPAs and HRGPs-extensin changes as an effect of cell wall rebuilding in two types of PVYNTN- Solanum tuberosum interactions [J]. Viruses, 2020, 12(1): 66. |
| 21 | Sampedro J, Cosgrove DJ. The expansin superfamily [J]. Genome Biol, 2005, 6(12): 242. |
| 22 | Ding AM, Marowa P, Kong YZ. Genome-wide identification of the expansin gene family in tobacco (Nicotiana tabacum) [J]. Mol Genet Genomics, 2016, 291(5): 1891-1907. |
| 23 | Lu YE, Liu LF, Wang X, et al. Genome-wide identification and expression analysis of the expansin gene family in tomato [J]. Mol Genet Genomics, 2016, 291(2): 597-608. |
| 24 | Hou L, Zhang ZY, Dou SH, et al. Genome-wide identification, characterization, and expression analysis of the expansin gene family in Chinese jujube (Ziziphus jujuba Mill.) [J]. Planta, 2019, 249(3): 815-829. |
| 25 | 廖嘉明. 基于CRISPR/Cas9的拟南芥EXPA多基因突变体的表现及转黄梁木NcEXPA8的表达 [D]. 广州: 华南农业大学, 2020. |
| Liao JM. Expression of EXPA multigene mutant in Arabidopsis thaliana based on CRISPR/Cas9 and expression of NcEXPA8 in Pistacia chinensis [D]. Guangzhou: South China Agricultural University, 2020. | |
| 26 | Sánchez-Montesino R, Bouza-Morcillo L, Marquez J, et al. A regulatory module controlling GA-mediated endosperm cell expansion is critical for seed germination in Arabidopsis [J]. Mol Plant, 2019, 12(1): 71-85. |
| 27 | Jiang XS, Li HY, Wang T, et al. Gibberellin indirectly promotes chloroplast biogenesis as a means to maintain the chloroplast population of expanded cells [J]. Plant J, 2012, 72(5): 768-780. |
| 28 | Park CH, Kim TW, Son SH, et al. Brassinosteroids control AtEXPA5 gene expression in Arabidopsis thaliana [J]. Phytochemistry, 2010, 71(4): 380-387. |
| 29 | Kuluev BR, Knyazev AB, Lebedev YP, et al. Morphological and physiological characteristics of transgenic tobacco plants expressing expansin genes: AtEXP10 from Arabidopsis and PnEXPA1 from poplar [J]. Russ J Plant Physiol, 2012, 59(1): 97-104. |
| 30 | Dermatsev V, Weingarten-Baror C, Resnick N, et al. Microarray analysis and functional tests suggest the involvement of expansins in the early stages of symbiosis of the arbuscular mycorrhizal fungus Glomus intraradices on tomato (Solanum lycopersicum) [J]. Mol Plant Pathol, 2010, 11(1): 121-135. |
| 31 | Gal TZ, Aussenberg ER, Burdman S, et al. Expression of a plant expansin is involved in the establishment of root knot nematode parasitism in tomato [J]. Planta, 2006, 224(1): 155-162. |
| 32 | Mohanty SK, Arthikala MK, Nanjareddy K, et al. Plant-symbiont interactions: the functional role of expansins [J]. Symbiosis, 2018, 74(1): 1-10. |
| 33 | Sasidharan R, Voesenek LA, Pierik R. Cell wall modifying proteins mediate plant acclimatization to biotic and abiotic stresses [J]. Crit Rev Plant Sci, 2011, 30(6): 548-562. |
| 34 | Hu HZ, Tang YW, Wu J, et al. Brassica napus mediator Subunit16 induces BnMED25- and BnWRKY33-activated defense signaling to confer Sclerotinia sclerotiorum resistance [J]. Front Plant Sci, 2021, 12: 663536. |
| 35 | Balbi V, Devoto A. Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios [J]. New Phytol, 2008, 177(2): 301-318. |
| 36 | 吴健, 周永明, 王幼平. 油菜与核盘菌互作分子机理研究进展 [J]. 中国油料作物学报, 2018, 40(5): 721-729. |
| Wu J, Zhou YM, Wang YP. Research progress on molecular mechanisms of Brassica napus-Sclerotinia sclerotiorum interaction [J]. Chin J Oil Crop Sci, 2018, 40(5): 721-729. | |
| 37 | Lou Y, Zhou HS, Han Y, et al. Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana [J]. New Phytol, 2018, 217(1): 378-391. |
| 38 | Zhang SY, Wang J, Chen GH, et al. Functional analysis of a MYB transcription factor BrTDF1 in the tapetum development of Wucai (Brassica rapa ssp.) [J]. Sci Hortic, 2019, 257: 108728. |
| 39 | 徐佳文. 棉花α-Expansin-like基因GhEXLA1的功能解析 [D]. 武汉: 华中农业大学, 2022. |
| Xu JW. Functional analysis of cotton α-expansin-like gene GhEXLA1 [D]. Wuhan: Huazhong Agricultural University, 2022. | |
| 40 | 梁绮文. 干旱胁迫下普通烟草类扩展蛋白NtEXLA2和NtEXLA4基因功能分析 [D]. 昆明: 云南农业大学, 2022. |
| Liang QW. Functional analysis of NtEXLA2 and NtEXLA4 genes of common tobacco under drought stress [D]. Kunming: Yunnan Agricultural University, 2022. |
| [1] | YANG Yong, YUAN Guo-mei, KANG Xiao-xiao, LIU Ya-ming, WANG Dong-sheng, ZHANG Hai-e. Identification and Expression Analysis of Members of the SWEET Gene Family in Chinese Chestnut [J]. Biotechnology Bulletin, 2025, 41(2): 257-269. |
| [2] | YANG Jia-bao, ZHOU Zhi-ming, ZHANG Zhan, FENG Li, SUN Li. Cloning,Expression of Helianthus annuus HaLACS1 Gene and Identification of Its Functional Complementation in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2022, 38(6): 147-156. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||