








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (6): 87-98.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1215
Previous Articles Next Articles
WU Hao1( ), DONG Wei-feng1, HE Zi-tian1, LI Yan-xiao1, XIE Hui2, SUN Ming-zhe1,3, SHEN Yang1,4(
), DONG Wei-feng1, HE Zi-tian1, LI Yan-xiao1, XIE Hui2, SUN Ming-zhe1,3, SHEN Yang1,4( ), SUN Xiao-li1(
), SUN Xiao-li1( )
)
Received:2024-12-17
Online:2025-06-26
Published:2025-04-23
Contact:
SHEN Yang, SUN Xiao-li
E-mail:2864288928@qq.com;shenyang2024@byau.edu.cn;csmbl2016@126.com
WU Hao, DONG Wei-feng, HE Zi-tian, LI Yan-xiao, XIE Hui, SUN Ming-zhe, SHEN Yang, SUN Xiao-li. Genome-wide Identification and Expression Analysis of the Rice BXL Gene Family[J]. Biotechnology Bulletin, 2025, 41(6): 87-98.
| 基因名称 Gene name | 上游引物 Forward primer (5'-3') | 下游引物 Reverse primer (5'-3') |
|---|---|---|
| OsBXL1 | GCTGACCAACCTCTACCTCAC | TGCCACCGAATTGACCTTCT |
| OsBXL2 | CACGTACAACCCTCCGTTCA | CCTCCAATCTCCCCTTGCAG |
| OsBXL3 | TACGGAGAGGAAGGCAGGAT | CTTGCACGACCTTCCTGGAT |
| OsBXL4 | AGCACTACACCGCATACGAC | AGCACATAACACTGGCCACA |
| OsBXL8 | AACGGTGTCCAACGCTACAA | GGCGCACATTATGCACGTAG |
| OsELf1-α | CATGATCACCGGTACCTCG | CCAGCATGTTGTCTCCGTG |
Table 1 Primers for quantitative fluorescence PCR
| 基因名称 Gene name | 上游引物 Forward primer (5'-3') | 下游引物 Reverse primer (5'-3') |
|---|---|---|
| OsBXL1 | GCTGACCAACCTCTACCTCAC | TGCCACCGAATTGACCTTCT |
| OsBXL2 | CACGTACAACCCTCCGTTCA | CCTCCAATCTCCCCTTGCAG |
| OsBXL3 | TACGGAGAGGAAGGCAGGAT | CTTGCACGACCTTCCTGGAT |
| OsBXL4 | AGCACTACACCGCATACGAC | AGCACATAACACTGGCCACA |
| OsBXL8 | AACGGTGTCCAACGCTACAA | GGCGCACATTATGCACGTAG |
| OsELf1-α | CATGATCACCGGTACCTCG | CCAGCATGTTGTCTCCGTG |

Fig. 1 Functional domains and sequence alignment of OsBXL proteinsA: Schematic diagram of structural domains in proteins. B: Multiple sequence alignment of proteins; SP: signal peptide, Glyco_hydro_3 domain: N-terminal catalytic domain of glycoside hydrolase; Glyco_hydro_3_C domain: C-terminal catalytic domain of glycoside hydrolase; Fn 3-like domain: fibronectin type Ⅲ domain. red boxes mark the WGR and KH conserved sites, the blue triangle refers to the nucleophilic catalytic amino acid residue site Asp, the black triangle refers to the acid-base catalytic amino acid residue Glu
基因名称 Gene name | 基因登录号 Gene ID | 蛋白长度 Length of protein sequences (aa) | 分子量 Molecular weight (kD) | 理论等电点 Theoretical pI | 亲疏水性 GRAVY | 亚细胞定位/置信度 Subcellular location/Probability | 信号肽位置 Signal peptide position (aa) |
|---|---|---|---|---|---|---|---|
| OsBXL1 | LOC_Os01g19220 | 771 | 82.60 | 6.61 | -0.191 | 细胞壁/0.7852 Cell wall | 1-24 |
| OsBXL2 | LOC_Os02g51620 | 818 | 87.12 | 5.21 | 0.030 | 细胞壁/0.7618 Cell wall | 1-27 |
| OsBXL3 | LOC_Os04g44840 | 782 | 83.43 | 5.77 | -0.110 | 细胞壁/0.753 Cell wall | 1-20 |
| OsBXL4 | LOC_Os04g54810 | 770 | 83.65 | 5.84 | -0.111 | 细胞壁/0.8309 Cell wall | 1-26 |
| OsBXL5 | LOC_Os11g18690 | 793 | 85.03 | 6.33 | -0.103 | 细胞壁/0.5974 Cell wall | 1-26 |
| OsBXL6 | LOC_Os11g18730 | 780 | 84.30 | 6.32 | -0.146 | 细胞壁/0.8285 Cell wall | 1-22 |
| OsBXL7 | LOC_Os11g19160 | 853 | 91.66 | 9.10 | -0.130 | 细胞壁/0.7545 Cell wall | 1-21 |
| OsBXL8 | LOC_Os11g19210 | 816 | 88.75 | 6.19 | -0.150 | 细胞壁/0.6741 Cell wall | 1-31 |
| OsBXL9 | LOC_Os11g44950 | 765 | 80.92 | 6.42 | 0.022 | 细胞壁/0.8602 Cell wall | 1-18 |
| OsBXL10 | LOC_Os11g47350 | 664 | 70.19 | 7.92 | -0.069 | 细胞壁/0.7416 Cell wall | 1-24 |
Table 2 Basic information of OsBXLs
基因名称 Gene name | 基因登录号 Gene ID | 蛋白长度 Length of protein sequences (aa) | 分子量 Molecular weight (kD) | 理论等电点 Theoretical pI | 亲疏水性 GRAVY | 亚细胞定位/置信度 Subcellular location/Probability | 信号肽位置 Signal peptide position (aa) |
|---|---|---|---|---|---|---|---|
| OsBXL1 | LOC_Os01g19220 | 771 | 82.60 | 6.61 | -0.191 | 细胞壁/0.7852 Cell wall | 1-24 |
| OsBXL2 | LOC_Os02g51620 | 818 | 87.12 | 5.21 | 0.030 | 细胞壁/0.7618 Cell wall | 1-27 |
| OsBXL3 | LOC_Os04g44840 | 782 | 83.43 | 5.77 | -0.110 | 细胞壁/0.753 Cell wall | 1-20 |
| OsBXL4 | LOC_Os04g54810 | 770 | 83.65 | 5.84 | -0.111 | 细胞壁/0.8309 Cell wall | 1-26 |
| OsBXL5 | LOC_Os11g18690 | 793 | 85.03 | 6.33 | -0.103 | 细胞壁/0.5974 Cell wall | 1-26 |
| OsBXL6 | LOC_Os11g18730 | 780 | 84.30 | 6.32 | -0.146 | 细胞壁/0.8285 Cell wall | 1-22 |
| OsBXL7 | LOC_Os11g19160 | 853 | 91.66 | 9.10 | -0.130 | 细胞壁/0.7545 Cell wall | 1-21 |
| OsBXL8 | LOC_Os11g19210 | 816 | 88.75 | 6.19 | -0.150 | 细胞壁/0.6741 Cell wall | 1-31 |
| OsBXL9 | LOC_Os11g44950 | 765 | 80.92 | 6.42 | 0.022 | 细胞壁/0.8602 Cell wall | 1-18 |
| OsBXL10 | LOC_Os11g47350 | 664 | 70.19 | 7.92 | -0.069 | 细胞壁/0.7416 Cell wall | 1-24 |

Fig. 2 Phylogenetic relation analysis of rice BXL familyThe green area indicates Group I, the light green area indicates Group II, and the orange area indicates Group Ⅲ

Fig. 3 Gene structures and conserved motifs of OsBXLsA: Phylogenetic tree of the OsBXL genefamily. B: The intron-exon architecture, yellow boxes indicate CDS, green boxes indicate UTR, and solid lines indicate intron. C: Conserved motifs, different colored boxes at the bottom indicate different motifs
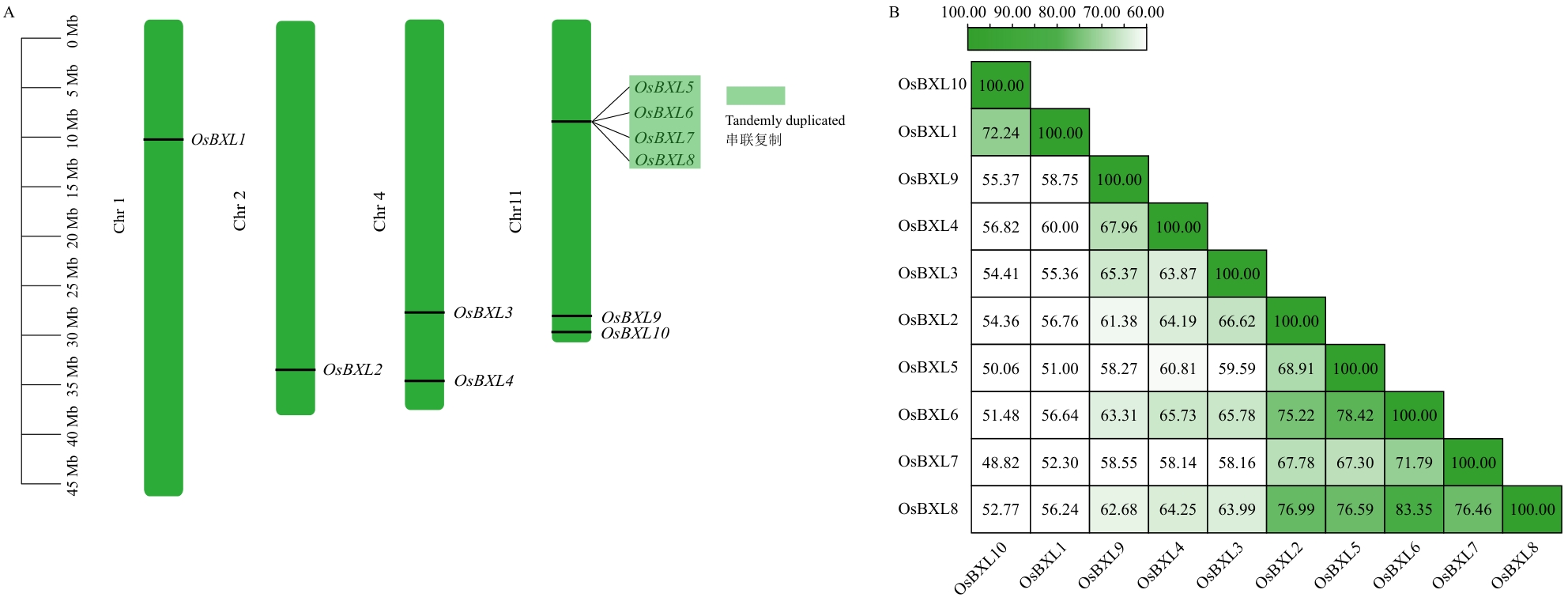
Fig. 4 Chromosomal location and protein sequence similarity of OsBXLsA: Chromosomal location, with the light green areas highlighting gene cluster containing four members. B: Protein sequence similarity of OsBXLs, with the numbers shown in the figure indicating amino acid similarity expressed as a percentage (%)
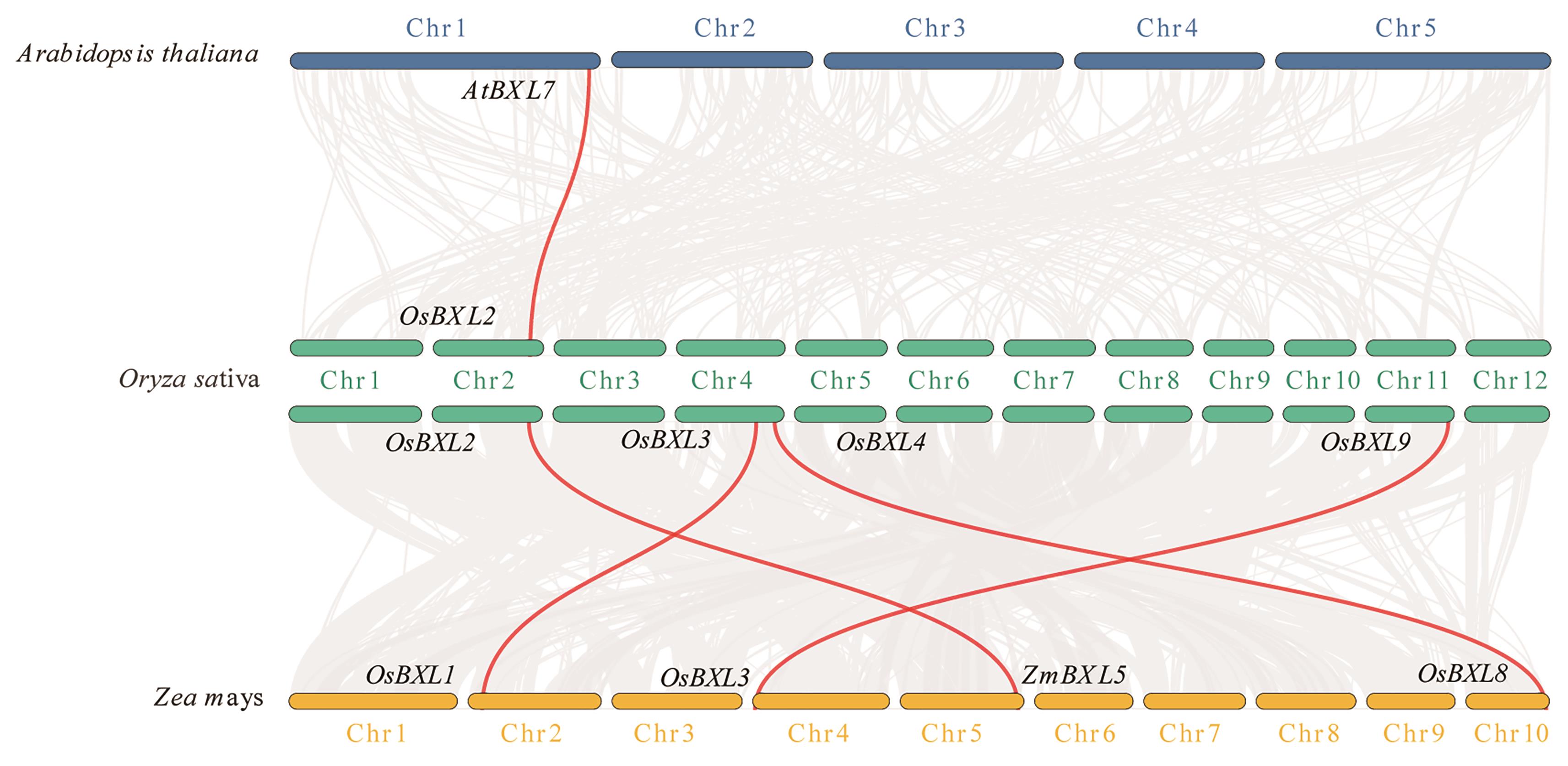
Fig. 5 Co-linearity analysis of BXL family genes in rice, maize, and ArabidopsisThe background gray lines indicate synteny in the genomes of rice with maize and Arabidopsis. The red lines indicate the synteny of OsBXLs with ZmBXLs and AtBXLs. The blue, green, and yellow bars indicate the chromosomes of Arabidopsis, rice, and maize, respectively

Fig. 6 Tissue expression pattern of OsBXLs genes family membersThe red line area corresponds to Cluster I, while the green line area corresponds to Cluster II; Log2FPKM >7 is defined as high expression, and Log2FPKM <7 is defined as low expression

Fig. 7 Haplotype analysis of OsBXL7 based on the coding sequenceA: Schematic of the base and amino acid variations in different haplotypes of OsBXL7, CDS: coding sequence; AA: amino acids; REF: reference haplotype. B: Distribution of varieties with different haplotypes of OsBXL7. C-F: The grain length, grain width, length/width ratio and thousand-grain weight of rice varieties with different haplotypes of OsBXL7, respectively. P values were calculated by one-way ANOVA analysis
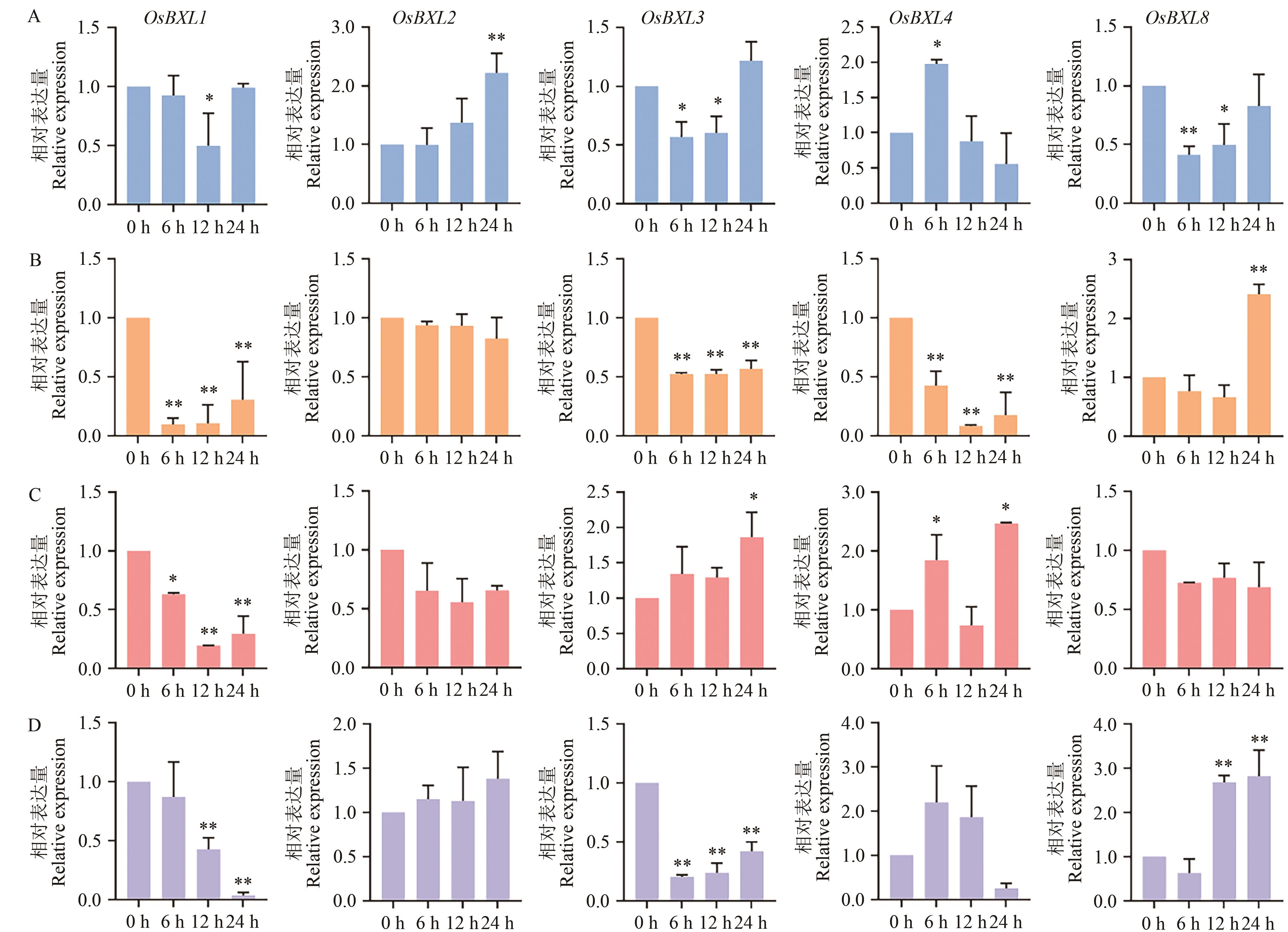
Fig. 8 Expression analysis of OsBXLs under abiotic stress and hormone treatmentA: Drought stress. B: Salt-alkali stress. C: ABA treatment. D: MeJA treatment; * and ** indicate P <0.05, P <0.01, respectively
| [1] | Weidenhamer JD, Cipollini D, Morris K, et al. Ecological realism and rigor in the study of plant-plant allelopathic interactions [J]. Plant Soil, 2023, 489(1): 1-39. |
| [2] | Pandey P, Ramegowda V, Senthil-Kumar M. Shared and unique responses of plants to multiple individual stresses and stress combinations: physiological and molecular mechanisms [J]. Front Plant Sci, 2015, 6: 723. |
| [3] | Wan JX, He M, Hou QQ, et al. Cell wall associated immunity in plants [J]. Stress Biol, 2021, 1(1): 3. |
| [4] | 张曼, 张叶卓, 何其邹洪, 等. 植物细胞壁结构及成像技术研究进展 [J]. 生物技术通报, 2023, 39(7): 113-122. |
| Zhang M, Zhang YZ, He QZH, et al. Advances in plant cell wall structure and imaging technology [J]. Biotechnol Bull, 2023, 39(7): 113-122. | |
| [5] | Shtein I, Bar-On B, Popper ZA. Plant and algal structure: from cell walls to biomechanical function [J]. Physiol Plant, 2018, 164(1): 56-66. |
| [6] | Cleemput G, Hessing M, Van Oort M, et al. Purification and characterization of a β-D-xylosidase and an endo-xylanase from wheat flour [J]. Plant Physiol, 1997, 113(2): 377-386. |
| [7] | Li N, Zhang R, Zhou JP, et al. Structures, biochemical characteristics, and functions of β-xylosidases [J]. J Agric Food Chem, 2023, 71(21): 7961-7976. |
| [8] | Pontonio E, Mahony J, Di Cagno R, et al. Cloning, expression and characterization of a β-D-xylosidase from Lactobacillus rossiae DSM 15814(T) [J]. Microb Cell Fact, 2016, 15: 72. |
| [9] | Knob A, Terrasan CRF, Carmona EC. β-xylosidases from filamentous fungi: an overview [J]. World J Microbiol Biotechnol, 2010, 26(3): 389-407. |
| [10] | Minic Z, Rihouey C, Do CT, et al. Purification and characterization of enzymes exhibiting beta-D-xylosidase activities in stem tissues of Arabidopsis [J]. Plant Physiol, 2004, 135(2): 867-878. |
| [11] | Goujon T, Minic Z, El Amrani A, et al. AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development [J]. Plant J, 2003, 33(4): 677-690. |
| [12] | Chen JY, Qu CP, Chang RH, et al. Genome-wide identification of BXL genes in Populus trichocarpa and their expression under different nitrogen treatments [J]. 3 Biotech, 2020, 10(2): 57. |
| [13] | Lee RC, Hrmova M, Burton RA, et al. Bifunctional family 3 glycoside hydrolases from barley with α-l-arabinofuranosidase and β-d-xylosidase activity characterization, primary structures, and cooh-terminal processing [J]. J Biol Chem, 2003, 278(7): 5377-5387. |
| [14] | 赖彪, 陈春帆, 刘春渝, 等. 茎瘤芥BXL基因家族的全基因组鉴定及在瘤状茎膨大过程中的表达分析 [J]. 华中农业大学学报, 2020, 39(6): 155-163. |
| Lai B, Chen CF, Liu CY, et al. Genome-wide identification of BXL family genes in Brassica juncea var. tumida and their expression during swelling of stem [J]. J Huazhong Agric Univ, 2020, 39(6): 155-163. | |
| [15] | Carolina Di Santo M, Ilina N, Pagano EA, et al. A Japanese plum α-l-arabinofuranosidase/β-D-xylosidase gene is developmentally regulated by alternative splicing [J]. Plant Sci, 2015, 231: 173-183. |
| [16] | Collins T, Gerday C, Feller G. Xylanases, xylanase families and extremophilic xylanases [J]. FEMS Microbiol Rev, 2005, 29(1): 3-23. |
| [17] | Itai A, Yoshida K, Tanabe K, et al. A-D-xylosidase-like gene is expressed during fruit ripening in Japanese pear (Pyrus pyrifolia Nakai) [J]. J Exp Bot, 1999, 50(335): 877-878. |
| [18] | Bustamante CA, Civello PM, Martínez GA. Cloning of the promoter region of β-xylosidase (FaXyl1) gene and effect of plant growth regulators on the expression of FaXyl1 in strawberry fruit [J]. Plant Sci, 2009, 177(1): 49-56. |
| [19] | 丁文家, 胡峻铭, 王嘉力. 水稻育种主要目标性状基因挖掘研究进展 [J]. 杂交水稻, 2023, 38(3): 1-19. |
| Ding WJ, Hu JM, Wang JL. Research progress on gene mining of main target traits in rice breeding [J]. Hybrid Rice, 2023, 38(3): 1-19. | |
| [20] | 赵海成, 李红宇, 郑桂萍, 等. 寒地水稻新品种垦粳8号的选育及栽培技术 [J]. 黑龙江农业科学, 2021(1): 165-168. |
| Zhao HC, Li HY, Zheng GP, et al. Breeding and cultivation techniques of a new rice variety kenjing No.8 in cold region [J]. Heilongjiang Agric Sci, 2021(1): 165-168. | |
| [21] | Jain M, Nijhawan A, Arora R, et al. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress [J]. Plant Physiol, 2007, 143(4): 1467-1483. |
| [22] | Drula E, Garron ML, Dogan S, et al. The carbohydrate-active enzyme database: functions and literature [J]. Nucleic Acids Res, 2022, 50(D1): D571-D577. |
| [23] | Wang FM, Longkumer T, Catausan SC, et al. Genome-wide association and gene validation studies for early root vigour to improve direct seeding of rice [J]. Plant Cell Environ, 2018, 41(12): 2731-2743. |
| [24] | Guzha A, McGee R, Scholz P, et al. Cell wall-localized BETA-XYLOSIDASE4 contributes to immunity of Arabidopsis against Botrytis cinerea [J]. Plant Physiol, 2022, 189(3): 1794-1813. |
| [25] | 冯凯月, 赵鑫焱, 李子妍, 等. 植物响应盐碱胁迫的分子机制研究进展 [J]. 生物技术通报, 2024, 40(10): 122-138. |
| Feng KY, Zhao XY, Li ZY, et al. Research progress on molecular mechanism of plant response to saline-alkali stress [J]. Biotech.bull, 2024, 40(10): 122-138. | |
| [26] | 吴占清, 陈威, 赵展, 等. 玉米GRAS基因家族的全基因组鉴定及生物信息学分析 [J]. 中国农业科技导报, 2024, 26(3): 15-25. |
| Wu ZQ, Chen W, Zhao Z, et al. Genome-wide identification and bioinformatics analysis of GRAS gene family in maize [J]. J Agric Sci Technol, 2024, 26(3): 15-25. | |
| [27] | Cui D, Zhou H, Ma XD, et al. Genomic insights on the contribution of introgressions from Xian/Indica to the genetic improvement of Geng/Japonica rice cultivars [J]. Plant Commun, 2022, 3(3): 100325. |
| [28] | Ji C, Xu LN, Li YJ, et al. The O2-ZmGRAS11 transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize [J]. Mol Plant, 2022, 15(3): 468-487. |
| [29] | Bauer K, Nayem S, Lehmann M, et al. β-D-XYLOSIDASE 4 modulates systemic immune signaling in Arabidopsis thaliana [J]. Front Plant Sci, 2023, 13: 1096800. |
| [30] | Yu L, Wilson LFL, Terrett OM, et al. Evolution of glucuronoxylan side chain variability in vascular plants and the compensatory adaptations of cell wall-degrading hydrolases [J]. New Phytol, 2024, 244(3): 1024-1040. |
| [31] | Wang LQ, Guo K, Li Y, et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice [J]. BMC Plant Biol, 2010, 10: 282. |
| [32] | Yu XZ, Fan WJ, Lin YJ, et al. Differential expression of the PAL gene family in rice seedlings exposed to chromium by microarray analysis [J]. Ecotoxicology, 2018, 27(3): 325-335. |
| [33] | Sun LX, Deng RL, Liu JW, et al. An overview of sucrose transporter (SUT) genes family in rice [J]. Mol Biol Rep, 2022, 49(6): 5685-5695. |
| [34] | Xia D, Zhou H, Liu RJ, et al. GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice [J]. Mol Plant, 2018, 11(5): 754-756. |
| [35] | Liu JF, Chen J, Zheng XM, et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice [J]. Nat Plants, 2017, 3: 17043. |
| [36] | Gao XY, Zhang JQ, Zhang XJ, et al. Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling [J]. Plant Cell, 2019, 31(5): 1077-1093. |
| [37] | Zhu XY, Gou YJ, Heng YQ, et al. Targeted manipulation of grain shape genes effectively improves outcrossing rate and hybrid seed production in rice [J]. Plant Biotechnol J, 2023, 21(2): 381-390. |
| [38] | Nagashima Y, Ma ZY, Liu XT, et al. Multiple quality control mechanisms in the ER and TGN determine subcellular dynamics and salt-stress tolerance function of KORRIGAN1 [J]. Plant Cell, 2020, 32(2): 470-485. |
| [1] | HUANG Dan, PENG Bing-yang, ZHANG Pan-pan, JIAO Yue, LYU Jia-bin. Identification of HD-Zip Gene Family in Camellia oleifera and Analysis of Its Expression under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(6): 191-207. |
| [2] | PENG Shao-zhi, WANG Deng-ke, ZHANG Xiang, DAI Xiong-ze, XU Hao, ZOU Xue-xiao. Cloning, Expression Characteristics and Functional Verification of the Pepper CaFD1 Gene [J]. Biotechnology Bulletin, 2025, 41(5): 153-164. |
| [3] | LIU Yuan-yuan, CHEN Xi-feng, QIAN Qian, GAO Zhen-yu. Advances in Molecular Mechanisms Regulating Panicle Development in Rice [J]. Biotechnology Bulletin, 2025, 41(5): 1-13. |
| [4] | LIU Yuan, ZHAO Ran, LU Zhen-fang, LI Rui-li. Research Progress in the Biological Metabolic Pathway and Functions of Plant Carotenoids [J]. Biotechnology Bulletin, 2025, 41(5): 23-31. |
| [5] | DU Liang-heng, TANG Huang-lei, ZHANG Zhi-guo. Map-based Cloning of Light-responsive Gene ELM1 in Rice [J]. Biotechnology Bulletin, 2025, 41(5): 82-89. |
| [6] | CHEN Xiao-jun, HUI Jian, MA Hong-wen, BAI Hai-Bo, ZHONG Nan, LI Jia-run, FAN Yun-fang. Creating Rice Gerplasm Resources OsALS Rsistant to Herbicide through Single Base Gene Editing Technology [J]. Biotechnology Bulletin, 2025, 41(4): 106-114. |
| [7] | ZHANG Yi-xuan, MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan. Genome-wide Identification and Expression Profiles of DIR Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(3): 123-136. |
| [8] | HAN Jiang-tao, ZHANG Shuai-bo, QIN Ya-rui, HAN Shuo-yang, ZHANG Ya-kang, WANG Ji-qing, DU Qing-jie, XIAO Huai-juan, LI Meng. Identification of β-amylase Gene Family in Melon and Their Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(3): 171-180. |
| [9] | LI Xin-peng, ZHANG Wu-han, ZHANG Li, SHU Fu, HE Qiang, GUO Yang, DENG Hua-feng, WANG Yue, SUN Ping-yong. Creation of Rice Mutant by Gamma-ray and Its Molecular Identification [J]. Biotechnology Bulletin, 2025, 41(3): 35-43. |
| [10] | KUANG Jian-hua, CHENG Zhi-peng, ZHAO Yong-jing, YANG Jie, CHEN Run-qiao, CHEN Long-qing, HU Hui-zhen. Expression Analysis of the GH3 Gene Family in Nelumbo nucifera underHormonal and Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(2): 221-233. |
| [11] | FANG Hui-min, GU Yi-shu, ZHANG Jing, ZHANG Long. Isolation and Physicochemical Properties Analysis of Starch from Rice Leaves [J]. Biotechnology Bulletin, 2025, 41(2): 51-57. |
| [12] | GE Shi-jie, LIU Yi-de, ZHANG Hua-dong, NING Qiang, ZHU Zhan-wang, WANG Shu-ping, LIU Yi-ke. Identification and Expression Analysis of Protein Disulfide Isomerase Gene Family in Wheat [J]. Biotechnology Bulletin, 2025, 41(2): 85-96. |
| [13] | YIN Yuan, CHENG Shuang, LIU Ding-hao, DENG Xiao-xia, LI Kai-yue, WANG Jing-hong, LIN Ji-xiang. Research Progress in Exogenous Hydrogen Peroxide(H2O2)Affecting Plant Growth and Physiological Metabolism under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(1): 1-13. |
| [14] | WU Zhi-jian, LIU Guang-yang, LIN Zhi-hao, SHENG Bin, CHEN Ge, XU Xiao-min, WANG Jun-wei, XU Dong-hui. Research Progress of Nano-regulation of Vegetable Seed Germination and Its Mechanism [J]. Biotechnology Bulletin, 2025, 41(1): 14-24. |
| [15] | LI Yu-xin, LI Miao, DU Xiao-fen, HAN Kang-ni, LIAN Shi-chao, WANG Jun. Identification and Expression Analysis of SiSAP Gene Family in Foxtail Millet(Setaria italica) [J]. Biotechnology Bulletin, 2025, 41(1): 143-156. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||