








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (5): 82-89.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1246
Previous Articles Next Articles
DU Liang-heng1,2( ), TANG Huang-lei2, ZHANG Zhi-guo2(
), TANG Huang-lei2, ZHANG Zhi-guo2( )
)
Received:2024-12-24
Online:2025-05-26
Published:2025-06-05
Contact:
ZHANG Zhi-guo
E-mail:du1150267215@163.com;zhangzhiguo@caas.cn
DU Liang-heng, TANG Huang-lei, ZHANG Zhi-guo. Map-based Cloning of Light-responsive Gene ELM1 in Rice[J]. Biotechnology Bulletin, 2025, 41(5): 82-89.
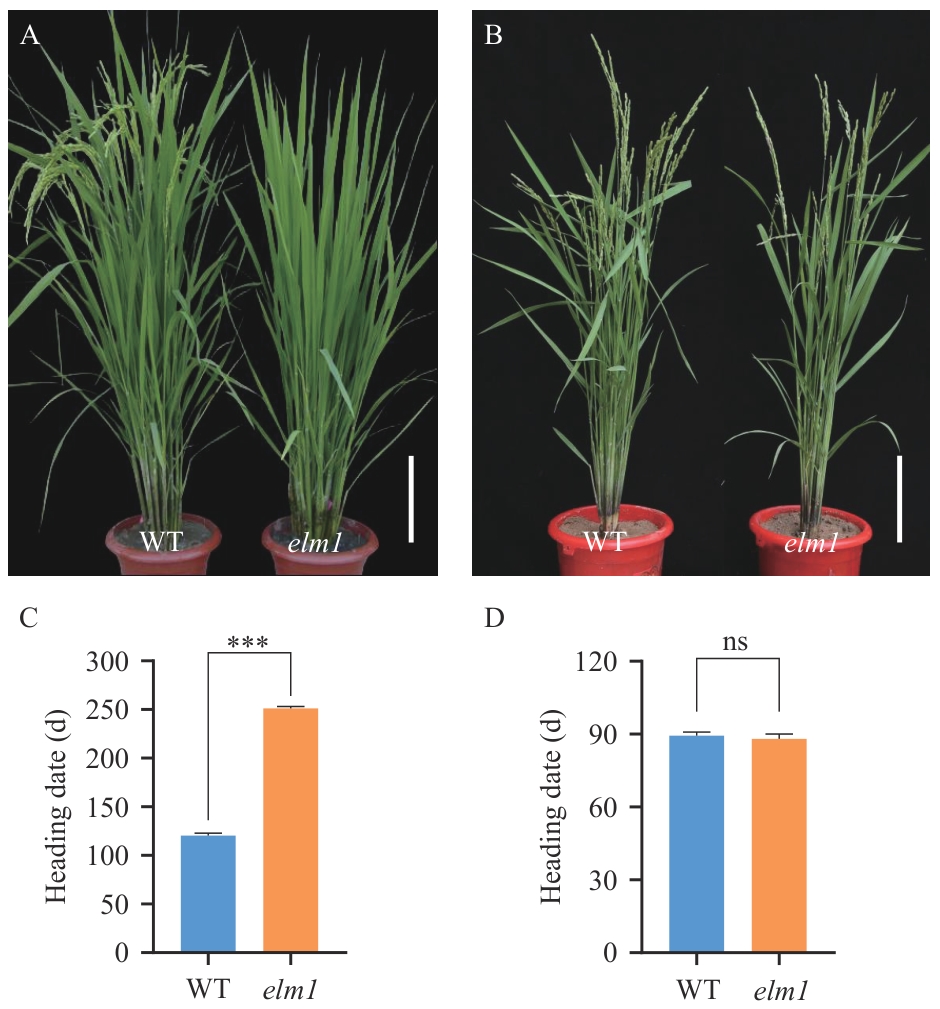
Fig. 1 Phenotypic of the flowering period between WT and elm1A: Comparison of wild type (WT) and elm1 mutant at the flowering stage in the experimental field in Beijing (Bar=20 cm). B: Comparison of WT and elm1 mutant at the flowering stage in Sanya, Hainan (Bar=20 cm). C: Comparison of heading date between WT and elm1 mutant in the experimental field in Beijing (long-day), *** indicates a significant difference at P<0.001. D: Comparison of heading date between WT and elm1 mutant in the experimental field in Sanya, Hainan (short-day), ns stands for not significant
性状类别 Trait category | 旗叶叶长 Flag leaf length (cm) | 旗叶叶宽 Flag leaf width (cm) | 株高 Plant height (cm) | 分蘖数 Tiller number | 千粒重 Thousand-grain weight (g) | 每穗粒数 Grain number per panicle |
|---|---|---|---|---|---|---|
| WT | 28.15±1.76a | 1.56±0.13a | 84.36±1.23a | 14±2a | 24.86±0.58a | 110±3a |
| elm1 | 29.56±1.13a | 1.47±0.17a | 80.58±2.16a | 11±3a | 25.13±0.67a | 120±2a |
Table 1 Comparison of agronomic traits between WT and elm1 mutant (Hainan)
性状类别 Trait category | 旗叶叶长 Flag leaf length (cm) | 旗叶叶宽 Flag leaf width (cm) | 株高 Plant height (cm) | 分蘖数 Tiller number | 千粒重 Thousand-grain weight (g) | 每穗粒数 Grain number per panicle |
|---|---|---|---|---|---|---|
| WT | 28.15±1.76a | 1.56±0.13a | 84.36±1.23a | 14±2a | 24.86±0.58a | 110±3a |
| elm1 | 29.56±1.13a | 1.47±0.17a | 80.58±2.16a | 11±3a | 25.13±0.67a | 120±2a |
统计类别 Statistical category | 日本晴× elm1 Nipponbare×elm1 | elm1×日本晴 elm1×Nipponbare |
|---|---|---|
植株总数 Total number of plants | 396 | 374 |
正常植株数 Number of normal plants | 288 | 276 |
不开花植株数 Number of non-flowering plants | 108 | 98 |
分离比 Segregation ratio | 3∶1 | 1∶3 |
| χ2 | 0.066 7 | 0.051 2 |
Table 2 Statistical results of F2 generation segregation population
统计类别 Statistical category | 日本晴× elm1 Nipponbare×elm1 | elm1×日本晴 elm1×Nipponbare |
|---|---|---|
植株总数 Total number of plants | 396 | 374 |
正常植株数 Number of normal plants | 288 | 276 |
不开花植株数 Number of non-flowering plants | 108 | 98 |
分离比 Segregation ratio | 3∶1 | 1∶3 |
| χ2 | 0.066 7 | 0.051 2 |
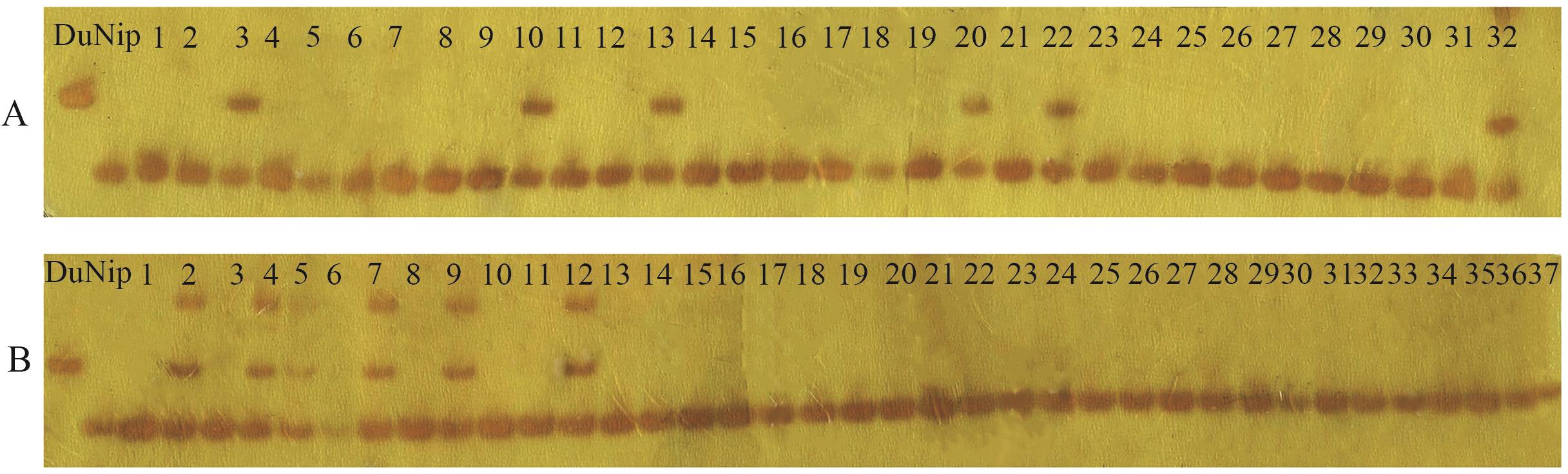
Fig. 2 Gel electrophoresis of gene linkage analysis for the mutant elm1A: Linkage analysis of the Indel-3 marker in individual F2 plants (1‒32: Electrophoresis banding results of 32 individual plants). B: Linkage analysis of the Indel-5 marker in individual F2 plants (1‒37: Electrophoresis banding results of 37 individual plants). Nip: Nipponbare. Du: Dular
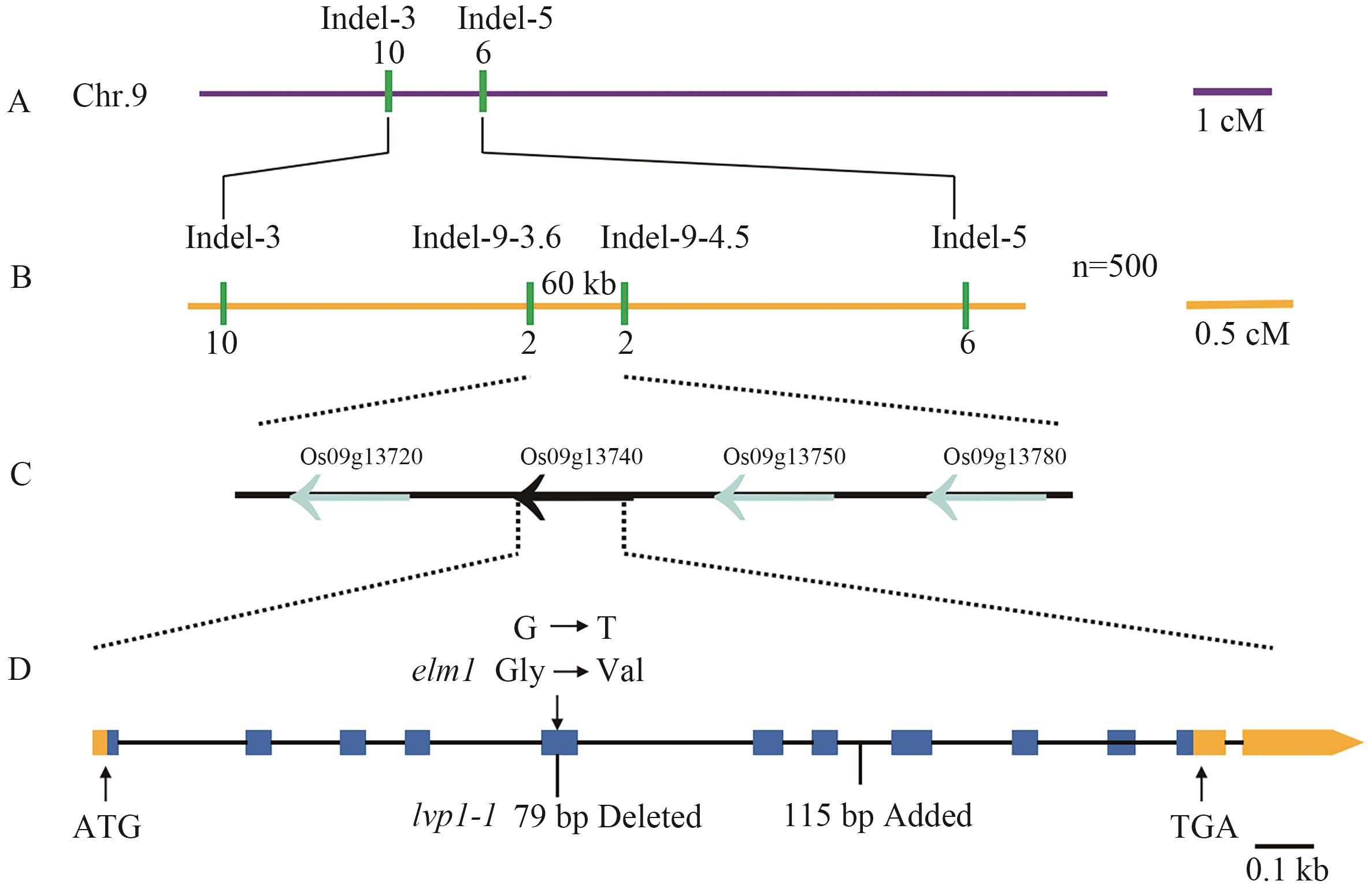
Fig. 3 Map-based cloning of gene ELM1A: Preliminary gene localization of elm1 mutant. The distance between each marker is represented by the recombination frequency, measured in centimorgans (cM). B: The fine mapping of the gene in the mutant elm1. C: Candidate region. D: The mutation in mutant elm1 occurs in the fifth exon of the LOC_Os09g13740, where the lvp1-1 mutation was a deletion of 79 base pairs in the fifth exon, and a mutation involving the insertion of 115 base pairs between the eighth and ninth exons

Fig. 4 Prediction of the ELM1 protein structure in WT and mutantA: Prediction of the ELM1 protein structure in the wild type. B: Prediction of the ELM1 protein structure after mutation
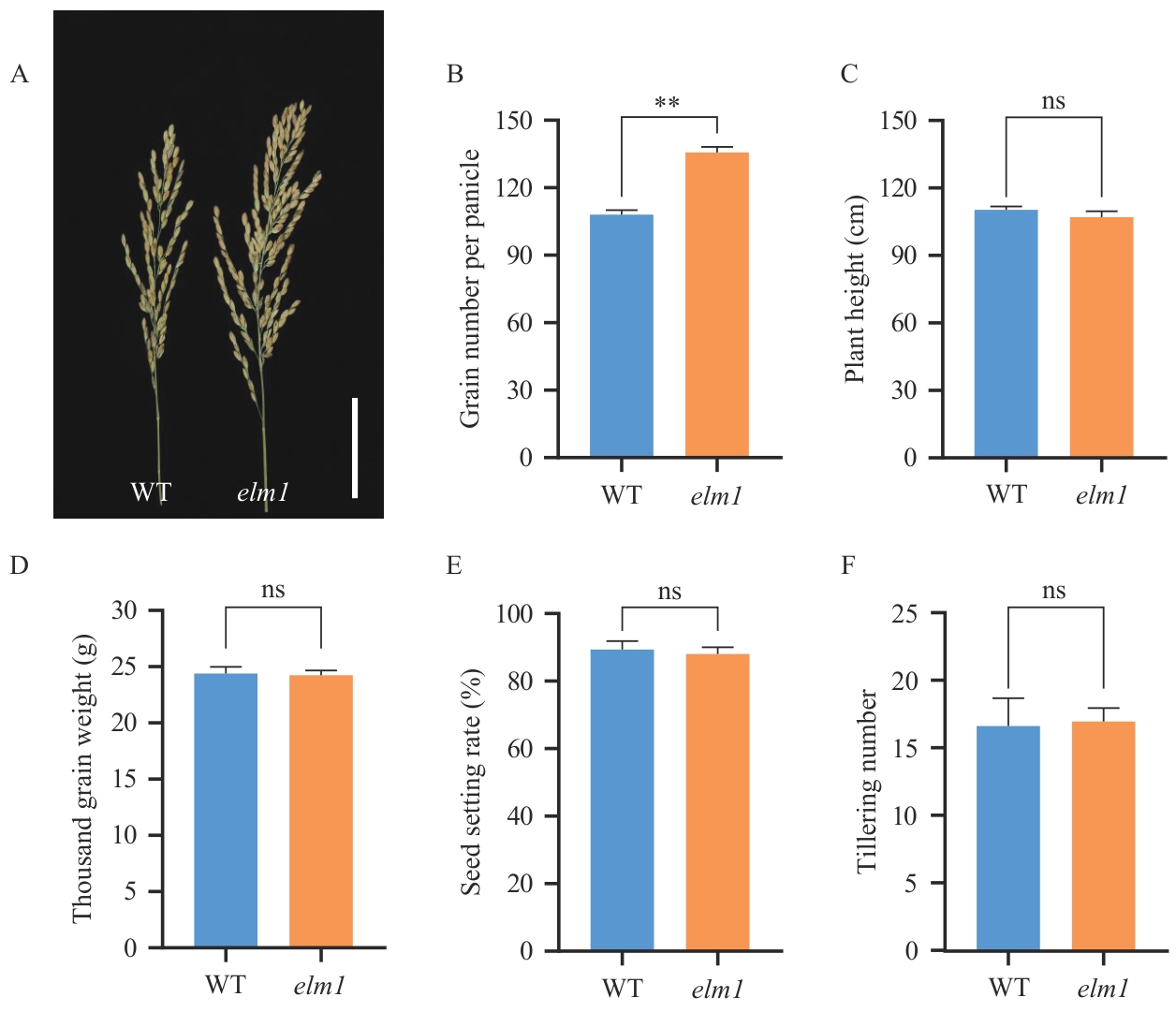
Fig. 5 Agronomic traits comparison between wild type and elm1 mutantA: Panicle comparison between WT and elm1 mutant (Bar=5 cm). B: Comparison of grain number per panicle between WT and elm1 mutant. ** indicates a significant difference at P<0.001. C: Comparison of plant height between WT and elm1 mutant. D: Comparison of thousand-grain weight between WT and elm1 mutant. E: Comparison of seed setting rate between WT and elm1 mutant. F: Comparison of tiller number between WT and elm1 mutant
| 1 | Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues [J]. Nat Rev Genet, 2012, 13(9): 627-639. |
| 2 | Song YH, Smith RW, To BJ, et al. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering [J]. Science, 2012, 336(6084): 1045-1049. |
| 3 | Wang JW. Regulation of flowering time by the miR156-mediated age pathway [J]. J Exp Bot, 2014, 65(17): 4723-4730. |
| 4 | Zhang SN, Zhang YY, Li KN, et al. Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice [J]. Curr Biol, 2021, 31(4): 671-683.e5. |
| 5 | Izawa T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice [J]. J Exp Bot, 2007, 58(12): 3091-3097. |
| 6 | Yano M, Katayose Y, Ashikari M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS [J]. Plant Cell, 2000, 12(12): 2473-2484. |
| 7 | Blümel M, Dally N, Jung C. Flowering time regulation in crops—what did we learn from Arabidopsis? [J]. Curr Opin Biotechnol, 2015, 32: 121-129. |
| 8 | Ga Z, Gao LY, Quzong XR, et al. Metabolomics, phytohormone and transcriptomics strategies to reveal the mechanism of barley heading date regulation to responds different photoperiod [J]. BMC Genomics, 2024, 25(1): 879. |
| 9 | Zhang ZY, Zhang B, Qi FX, et al. Hd1 function conversion in regulating heading is dependent on gene combinations of Ghd7, Ghd8, and Ghd7.1 under long-day conditions in rice [J]. Mol Breed, 2019, 39(7): 92. |
| 10 | Cui YX, Wang JR, Feng L, et al. A combination of long-day suppressor genes contributes to the northward expansion of rice [J]. Front Plant Sci, 2020, 11: 864. |
| 11 | Brambilla V, Fornara F. Y flowering? Regulation and activity of CONSTANS and CCT-domain proteins in Arabidopsis and crop species [J]. Biochim Biophys Acta Gene Regul Mech, 2017, 1860(5): 655-660. |
| 12 | Song YH, Shim JS, Kinmonth-Schultz HA, et al. Photoperiodic flowering: time measurement mechanisms in leaves [J]. Annu Rev Plant Biol, 2015, 66: 441-464. |
| 13 | Liu B, Liu YH, Wang BH, et al. The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment [J]. Nat Commun, 2019, 10(1): 2999. |
| 14 | Sun CH, Fang J, Zhao TL, et al. The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice [J]. Plant Cell, 2012, 24(8): 3235-3247. |
| 15 | Zong WB, Ren D, Huang MH, et al. Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1, Ghd7 and DTH8 in rice heading [J]. New Phytol, 2021, 229(3): 1635-1649. |
| 16 | Komiya R, Ikegami A, Tamaki S, et al. Hd3a and RFT1 are essential for flowering in rice [J]. Development, 2008, 135(4): 767-774. |
| 17 | Zheng QQ, Zhou ZJ, Li XR, et al. Heading date 3a stimulates tiller bud outgrowth in Oryza sativa L. through strigolactone signaling pathway [J]. Int J Mol Sci, 2024, 25(19): 10778. |
| 18 | Gao H, Zheng XM, Fei GL, et al. Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice [J]. PLoS Genet, 2013, 9(2): e1003281. |
| 19 | Zhang XN, Feng Q, Miao JS, et al. The WD40 domain-containing protein Ehd5 positively regulates flowering in rice (Oryza sativa) [J]. Plant Cell, 2023, 35(11): 4002-4019. |
| 20 | Wan SY, Wu JX, Zhang ZG, et al. Activation tagging, an efficient tool for functional analysis of the rice genome [J]. Plant Mol Biol, 2009, 69(1/2): 69-80. |
| 21 | Xue WY, Xing YZ, Weng XY, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice [J]. Nat Genet, 2008, 40(6): 761-767. |
| 22 | Ishimaru T, Hlaing KT, Oo YM, et al. An early-morning flowering trait in rice can enhance grain yield under heat stress field conditions at flowering stage [J]. Field Crops Res, 2022, 277: 108400. |
| 23 | Zhou SR, Zhu SS, Cui S, et al. Transcriptional and post-transcriptional regulation of heading date in rice [J]. New Phytol, 2021, 230(3): 943-956. |
| 24 | Yan WH, Wang P, Chen HX, et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice [J]. Mol Plant, 2011, 4(2): 319-330. |
| 25 | Wei XJ, Xu JF, Guo HN, et al. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously [J]. Plant Physiol, 2010, 153(4): 1747-1758. |
| 26 | Brambilla V, Gomez-Ariza J, Cerise M, et al. The importance of being on time: regulatory networks controlling photoperiodic flowering in cereals [J]. Front Plant Sci, 2017, 8: 665. |
| 27 | Jung C, Müller AE. Flowering time control and applications in plant breeding [J]. Trends Plant Sci, 2009, 14(10): 563-573. |
| 28 | Song J, Tang LQ, Cui YT, et al. Research progress on photoperiod gene regulation of heading date in rice [J]. Curr Issues Mol Biol, 2024, 46(9): 10299-10311. |
| 29 | Bai XF, Huang Y, Hu Y, et al. Duplication of an upstream silencer of FZP increases grain yield in rice [J]. Nat Plants, 2017, 3(11): 885-893. |
| 30 | Liu L, Gallagher J, Arevalo ED, et al. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes [J]. Nat Plants, 2021, 7(3): 287-294. |
| [1] | CHEN Xiao-jun, HUI Jian, MA Hong-wen, BAI Hai-Bo, ZHONG Nan, LI Jia-run, FAN Yun-fang. Creating Rice Gerplasm Resources OsALS Rsistant to Herbicide through Single Base Gene Editing Technology [J]. Biotechnology Bulletin, 2025, 41(4): 106-114. |
| [2] | LI Xin-peng, ZHANG Wu-han, ZHANG Li, SHU Fu, HE Qiang, GUO Yang, DENG Hua-feng, WANG Yue, SUN Ping-yong. Creation of Rice Mutant by Gamma-ray and Its Molecular Identification [J]. Biotechnology Bulletin, 2025, 41(3): 35-43. |
| [3] | FANG Hui-min, GU Yi-shu, ZHANG Jing, ZHANG Long. Isolation and Physicochemical Properties Analysis of Starch from Rice Leaves [J]. Biotechnology Bulletin, 2025, 41(2): 51-57. |
| [4] | JIN Su-kui, GUO Qian-qian, LIU Qiao-quan, GAO Ji-ping. A Simplified Method for Extracting Genomic DNA from Rice Leaves [J]. Biotechnology Bulletin, 2025, 41(1): 74-84. |
| [5] | LIU Wen-zhi, HE Dan, LI Peng, FU Ying-lin, ZHANG Yi-xin, WEN Hua-jie, YU Wen-qing. Paenibacillus polymyxa New Strain X-11 and Its Growth-promoting Effects on Tomato and Rice [J]. Biotechnology Bulletin, 2024, 40(9): 249-259. |
| [6] | LI Qing-mao, PENG Cong-gui, QI Xiao-han, LIU Xing-lei, LI Zhen-yuan, LI Qin-yan, HUANG Li-yu. Screening and Identification of Excellent Strains of Endophytic Bacteria Promoting Rice Iron Absorption from Wild Rice [J]. Biotechnology Bulletin, 2024, 40(8): 255-263. |
| [7] | SUN Zhi-yong, DU Huai-dong, LIU Yang, MA Jia-xin, YU Xue-ran, MA Wei, YAO Xin-jie, WANG Min, LI Pei-fu. Genome-wide Association Analysis of γ-aminobutyric Acid in Rice Grains [J]. Biotechnology Bulletin, 2024, 40(8): 53-62. |
| [8] | PANG Meng-zhen, XU Han-qin, LIU Hai-yan, SONG Juan, WANG Jia-han, SUN Li-na, JI Pei-mei, YIN Ze-zhi, HU You-chuan, ZHAO Xiao-meng, LIANG Shan-shan, ZHANG Si-ju, LUAN Wei-jiang. Gene Identification and Functional Analysis of Yellowish and Early Heading Mutant hz1 in Rice [J]. Biotechnology Bulletin, 2024, 40(7): 125-136. |
| [9] | TIAN Sheng-ni, ZHANG Qin, DONG Yu-fei, DING Zhou, YE Ai-hua, ZHANG Ming-zhu. Effects of Acid Mine Drainage on Physicochemical Factors and Nitrogen-fixing Microorganisms in the Root Zone of Mature Rice [J]. Biotechnology Bulletin, 2024, 40(6): 271-280. |
| [10] | KONG De-ting, QI Xiao-han, LIU Xing-lei, LI Li-ping, HU Feng-yi, HUANG Li-yu, QIN Shi-wen. Comparison and Analysis of Endophytic Bacterial Communities in Different Perennial Rice Varieties [J]. Biotechnology Bulletin, 2024, 40(5): 225-236. |
| [11] | DU Bing-shuai, ZOU Xin-hui, WANG Zi-hao, ZHANG Xin-yuan, CAO Yi-bo, ZHANG Ling-yun. Genome-wide Identification and Expression Analysis of the SWEET Gene Family in Camellia oleifera [J]. Biotechnology Bulletin, 2024, 40(5): 179-190. |
| [12] | PENG Feng, YU Hai-xia, ZHANG Kun, LIU Ying-ying, TAN Gui-yu. Review on the Regulation of Caleosin on Plant Lipid Droplet [J]. Biotechnology Bulletin, 2024, 40(4): 33-39. |
| [13] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [14] | LI Xing-rong, TAN Zhi-bing, ZHAO Yan, LI Yao-kui, ZHAO Bing-ran, TANG Li. Cloning and Functional Analysis of OsLCT3, a Low-affinity Cation Transporter Gene of Rice [J]. Biotechnology Bulletin, 2024, 40(4): 97-109. |
| [15] | LIU Jia-ning, LI Meng, YANG Xin-sen, WU Wei, PEI Xin-wu, YUAN Qian-hua. Impact of Different Water Management Cultivation Methods on the Rhizosphere Bacteria Community of Shanlan Upland Rice [J]. Biotechnology Bulletin, 2024, 40(3): 242-250. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||