








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (3): 123-136.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0825
ZHANG Yi-xuan( ), MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan(
), MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan( )
)
Received:2024-08-25
Online:2025-03-26
Published:2025-03-20
Contact:
HOU Hua-lan
E-mail:15045930755@stu.ahau.edu.cn;hhl@ahau.edu.cn
ZHANG Yi-xuan, MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan. Genome-wide Identification and Expression Profiles of DIR Gene Family in Potato[J]. Biotechnology Bulletin, 2025, 41(3): 123-136.
| 基因名Gene name | 基因ID Gene ID | 蛋白长度Length of protein/aa | 分子量Molecular weight/kD | 等电点pI | 不稳定系数Instability index | 脂肪系数Aliphatic index | 亲水性平均值Grand average of hydropathicity | 信号肽Signal peptide/aa | N-糖基化位点N-Glyc position | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|---|
| StDIR1 | Soltu.DM.01G001610.1 | 192 | 21.26 | 9.76 | 31.2 | 100.47 | 0.167 | 1-24 | 41 | Chloroplast |
| StDIR2 | Soltu.DM.01G012660.1 | 189 | 21.17 | 9.57 | 28 | 86.19 | 0.155 | 1-24 | - | Cell membrane. Chloroplast |
| StDIR3 | Soltu.DM.01G012790.1 | 191 | 21.29 | 9.37 | 24.95 | 77.12 | -0.019 | 1-24 | - | Cell wall. Chloroplast Mitochondrion |
| StDIR4 | Soltu.DM.01G019610.1 | 174 | 19.40 | 6.5 | 10.61 | 99.71 | 0.21 | - | - | Cell membrane |
| StDIR5 | Soltu.DM.01G050810.1 | 127 | 13.56 | 5.6 | 9.14 | 92.13 | 0.231 | 1-20 | - | Cell wall |
| StDIR6 | Soltu.DM.01G050820.1 | 188 | 20.76 | 9.1 | 31.15 | 90.27 | 0.062 | 1-22 | - | Cell membrane. Cell wall |
| StDIR7 | Soltu.DM.02G003630.1 | 190 | 21.10 | 8.76 | 27.85 | 92.37 | -0.015 | 1-22 | - | Cell membrane |
| StDIR8 | Soltu.DM.02G003640.1 | 185 | 20.57 | 9.1 | 25.39 | 91.19 | 0.001 | 1-17 | - | Cell membrane |
| StDIR9 | Soltu.DM.02G022680.1 | 318 | 34.60 | 9.56 | 27.65 | 91.01 | 0.041 | - | 289 315 | Endoplasmic reticulum |
| StDIR10 | Soltu.DM.04G003460.1 | 185 | 20.66 | 9.58 | 32.65 | 101.84 | 0.038 | - | 25 | Cell membrane |
| StDIR11 | Soltu.DM.04G007230.1 | 335 | 35.58 | 5.17 | 35.6 | 83.52 | -0.05 | 1-30 | - | Cell membrane |
| StDIR12 | Soltu.DM.05G023520.1 | 137 | 14.84 | 9.45 | 29.19 | 95.33 | 0.299 | 1-28 | - | Chloroplast. Peroxisome |
| StDIR13 | Soltu.DM.05G025340.1 | 400 | 41.21 | 4.29 | 41.77 | 88.55 | 0.023 | 1-30 | - | Cell membrane |
| StDIR14 | Soltu.DM.05G025350.1 | 240 | 26.46 | 6.34 | 34.48 | 78.38 | -0.238 | 1-25 | - | Nucleus |
| StDIR15 | Soltu.DM.06G015540.1 | 188 | 20.75 | 6.13 | 46.29 | 78.4 | 0.116 | 1-25 | - | Cell membrane |
| StDIR16 | Soltu.DM.06G031100.1 | 329 | 33.10 | 4.66 | 24.04 | 86.69 | 0.235 | 1-30 | - | Cell membrane |
| StDIR17 | Soltu.DM.07G013170.1 | 187 | 20.55 | 9.49 | 22.41 | 93.8 | 0.195 | 1-22 | - | Chloroplast |
| StDIR18 | Soltu.DM.07G017590.1 | 177 | 19.60 | 6.36 | 19.62 | 103.05 | 0.193 | 1-22 | 127 | Cell membrane Mitochondrion |
| StDIR19 | Soltu.DM.08G028530.1 | 189 | 21.53 | 6.35 | 25.21 | 70.69 | -0.086 | 1-24 | - | Cell membrane |
| StDIR20 | Soltu.DM.08G028540.1 | 189 | 21.16 | 6.9 | 20.34 | 91.9 | 0.1 | 1-26 | - | Cell membrane. Cell wall. Chloroplast |
| StDIR21 | Soltu.DM.09G028090.1 | 246 | 25.71 | 5.02 | 31.88 | 87.24 | 0.178 | 1-17 | 145 222 | Cell membrane |
| StDIR22 | Soltu.DM.10G004800.1 | 190 | 21.33 | 9.73 | 33.17 | 87.74 | 0.017 | 1-22 | 26 | Cell membrane |
| StDIR23 | Soltu.DM.10G017030.1 | 184 | 20.24 | 9.21 | 23.79 | 96.9 | 0.188 | 1-20 | 59 | Cell membrane. Cell wall. Chloroplast. Mitochondrion |
| StDIR24 | Soltu.DM.10G017040.1 | 194 | 20.93 | 9.27 | 20.18 | 101.6 | 0.176 | 1-26 | - | Cell membrane. Cell wall. Chloroplast |
| StDIR25 | Soltu.DM.10G017050.1 | 128 | 14.03 | 9.7 | 14.52 | 75.47 | 0.008 | 1-22 | 128 | Cell membrane |
| StDIR26 | Soltu.DM.10G017060.1 | 173 | 19.23 | 5.45 | 22.87 | 77.23 | -0.086 | - | - | Cell wall |
| StDIR27 | Soltu.DM.10G017070.1 | 194 | 20.90 | 9.3 | 17.68 | 105.62 | 0.213 | 1-26 | - | Cell membrane |
| StDIR28 | Soltu.DM.10G017090.1 | 193 | 21.18 | 9.52 | 16.09 | 82.28 | 0.069 | 1-23 | 75 | Cell membrane |
| StDIR29 | Soltu.DM.10G017100.1 | 174 | 19.35 | 5.81 | 20.61 | 79.02 | -0.113 | - | - | Cell wall |
| StDIR30 | Soltu.DM.10G017110.1 | 190 | 20.95 | 8.63 | 18.93 | 89.16 | 0.112 | 1-20 | 72 | Cell membrane. Cell wall |
| StDIR31 | Soltu.DM.10G017130.1 | 124 | 13.52 | 9.43 | 22.84 | 80.16 | 0.198 | - | - | Cell membrane |
| StDIR32 | Soltu.DM.10G017170.1 | 184 | 20.29 | 9.63 | 28.12 | 92.17 | 0.167 | 1-20 | - | Cell membrane. Cell wall |
| StDIR33 | Soltu.DM.11G026480.1 | 336 | 34.33 | 4.74 | 33.36 | 84.49 | 0.136 | 1-31 | 186 | Cell membrane |
| StDIR34 | Soltu.DM.12G003100.1 | 247 | 25.75 | 5.26 | 29.03 | 91.62 | 0.225 | 1-25 | 146 223 | Cell membrane |
Table 1 Sequence analysis of DIRs in potato
| 基因名Gene name | 基因ID Gene ID | 蛋白长度Length of protein/aa | 分子量Molecular weight/kD | 等电点pI | 不稳定系数Instability index | 脂肪系数Aliphatic index | 亲水性平均值Grand average of hydropathicity | 信号肽Signal peptide/aa | N-糖基化位点N-Glyc position | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|---|
| StDIR1 | Soltu.DM.01G001610.1 | 192 | 21.26 | 9.76 | 31.2 | 100.47 | 0.167 | 1-24 | 41 | Chloroplast |
| StDIR2 | Soltu.DM.01G012660.1 | 189 | 21.17 | 9.57 | 28 | 86.19 | 0.155 | 1-24 | - | Cell membrane. Chloroplast |
| StDIR3 | Soltu.DM.01G012790.1 | 191 | 21.29 | 9.37 | 24.95 | 77.12 | -0.019 | 1-24 | - | Cell wall. Chloroplast Mitochondrion |
| StDIR4 | Soltu.DM.01G019610.1 | 174 | 19.40 | 6.5 | 10.61 | 99.71 | 0.21 | - | - | Cell membrane |
| StDIR5 | Soltu.DM.01G050810.1 | 127 | 13.56 | 5.6 | 9.14 | 92.13 | 0.231 | 1-20 | - | Cell wall |
| StDIR6 | Soltu.DM.01G050820.1 | 188 | 20.76 | 9.1 | 31.15 | 90.27 | 0.062 | 1-22 | - | Cell membrane. Cell wall |
| StDIR7 | Soltu.DM.02G003630.1 | 190 | 21.10 | 8.76 | 27.85 | 92.37 | -0.015 | 1-22 | - | Cell membrane |
| StDIR8 | Soltu.DM.02G003640.1 | 185 | 20.57 | 9.1 | 25.39 | 91.19 | 0.001 | 1-17 | - | Cell membrane |
| StDIR9 | Soltu.DM.02G022680.1 | 318 | 34.60 | 9.56 | 27.65 | 91.01 | 0.041 | - | 289 315 | Endoplasmic reticulum |
| StDIR10 | Soltu.DM.04G003460.1 | 185 | 20.66 | 9.58 | 32.65 | 101.84 | 0.038 | - | 25 | Cell membrane |
| StDIR11 | Soltu.DM.04G007230.1 | 335 | 35.58 | 5.17 | 35.6 | 83.52 | -0.05 | 1-30 | - | Cell membrane |
| StDIR12 | Soltu.DM.05G023520.1 | 137 | 14.84 | 9.45 | 29.19 | 95.33 | 0.299 | 1-28 | - | Chloroplast. Peroxisome |
| StDIR13 | Soltu.DM.05G025340.1 | 400 | 41.21 | 4.29 | 41.77 | 88.55 | 0.023 | 1-30 | - | Cell membrane |
| StDIR14 | Soltu.DM.05G025350.1 | 240 | 26.46 | 6.34 | 34.48 | 78.38 | -0.238 | 1-25 | - | Nucleus |
| StDIR15 | Soltu.DM.06G015540.1 | 188 | 20.75 | 6.13 | 46.29 | 78.4 | 0.116 | 1-25 | - | Cell membrane |
| StDIR16 | Soltu.DM.06G031100.1 | 329 | 33.10 | 4.66 | 24.04 | 86.69 | 0.235 | 1-30 | - | Cell membrane |
| StDIR17 | Soltu.DM.07G013170.1 | 187 | 20.55 | 9.49 | 22.41 | 93.8 | 0.195 | 1-22 | - | Chloroplast |
| StDIR18 | Soltu.DM.07G017590.1 | 177 | 19.60 | 6.36 | 19.62 | 103.05 | 0.193 | 1-22 | 127 | Cell membrane Mitochondrion |
| StDIR19 | Soltu.DM.08G028530.1 | 189 | 21.53 | 6.35 | 25.21 | 70.69 | -0.086 | 1-24 | - | Cell membrane |
| StDIR20 | Soltu.DM.08G028540.1 | 189 | 21.16 | 6.9 | 20.34 | 91.9 | 0.1 | 1-26 | - | Cell membrane. Cell wall. Chloroplast |
| StDIR21 | Soltu.DM.09G028090.1 | 246 | 25.71 | 5.02 | 31.88 | 87.24 | 0.178 | 1-17 | 145 222 | Cell membrane |
| StDIR22 | Soltu.DM.10G004800.1 | 190 | 21.33 | 9.73 | 33.17 | 87.74 | 0.017 | 1-22 | 26 | Cell membrane |
| StDIR23 | Soltu.DM.10G017030.1 | 184 | 20.24 | 9.21 | 23.79 | 96.9 | 0.188 | 1-20 | 59 | Cell membrane. Cell wall. Chloroplast. Mitochondrion |
| StDIR24 | Soltu.DM.10G017040.1 | 194 | 20.93 | 9.27 | 20.18 | 101.6 | 0.176 | 1-26 | - | Cell membrane. Cell wall. Chloroplast |
| StDIR25 | Soltu.DM.10G017050.1 | 128 | 14.03 | 9.7 | 14.52 | 75.47 | 0.008 | 1-22 | 128 | Cell membrane |
| StDIR26 | Soltu.DM.10G017060.1 | 173 | 19.23 | 5.45 | 22.87 | 77.23 | -0.086 | - | - | Cell wall |
| StDIR27 | Soltu.DM.10G017070.1 | 194 | 20.90 | 9.3 | 17.68 | 105.62 | 0.213 | 1-26 | - | Cell membrane |
| StDIR28 | Soltu.DM.10G017090.1 | 193 | 21.18 | 9.52 | 16.09 | 82.28 | 0.069 | 1-23 | 75 | Cell membrane |
| StDIR29 | Soltu.DM.10G017100.1 | 174 | 19.35 | 5.81 | 20.61 | 79.02 | -0.113 | - | - | Cell wall |
| StDIR30 | Soltu.DM.10G017110.1 | 190 | 20.95 | 8.63 | 18.93 | 89.16 | 0.112 | 1-20 | 72 | Cell membrane. Cell wall |
| StDIR31 | Soltu.DM.10G017130.1 | 124 | 13.52 | 9.43 | 22.84 | 80.16 | 0.198 | - | - | Cell membrane |
| StDIR32 | Soltu.DM.10G017170.1 | 184 | 20.29 | 9.63 | 28.12 | 92.17 | 0.167 | 1-20 | - | Cell membrane. Cell wall |
| StDIR33 | Soltu.DM.11G026480.1 | 336 | 34.33 | 4.74 | 33.36 | 84.49 | 0.136 | 1-31 | 186 | Cell membrane |
| StDIR34 | Soltu.DM.12G003100.1 | 247 | 25.75 | 5.26 | 29.03 | 91.62 | 0.225 | 1-25 | 146 223 | Cell membrane |
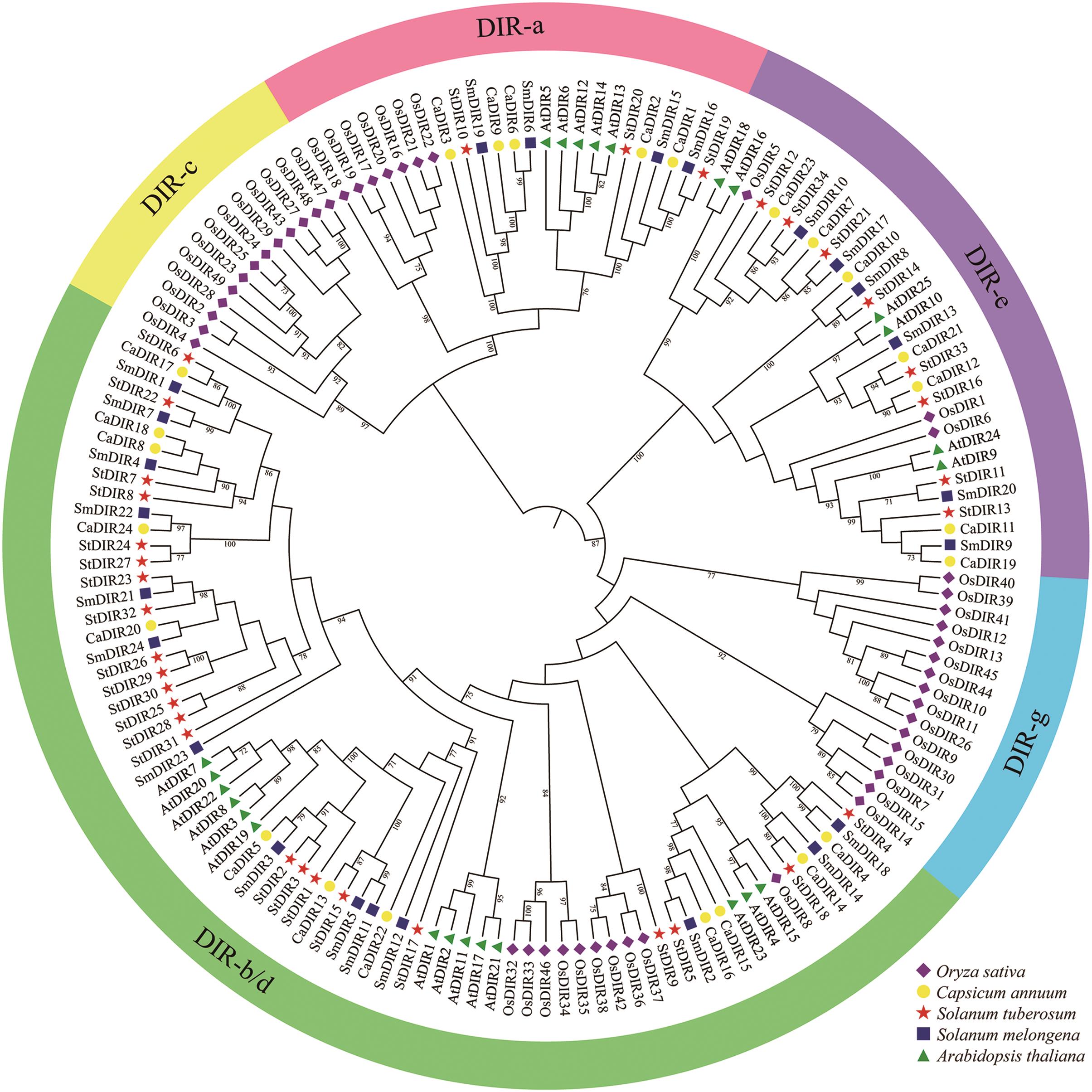
Fig. 2 Phylogenetic relationship of StDIRs from potato and DIRs from other plant speciesDifferent groups of DIRs are represented by different colors. Different icons indicate different species, O. sativa (Os), C. annuum (Ca), S. tuberosum (St), S. melongena (Sm), and A. thaliana (At) is indicated by rhombus, circle, pentagram, square, and triangle
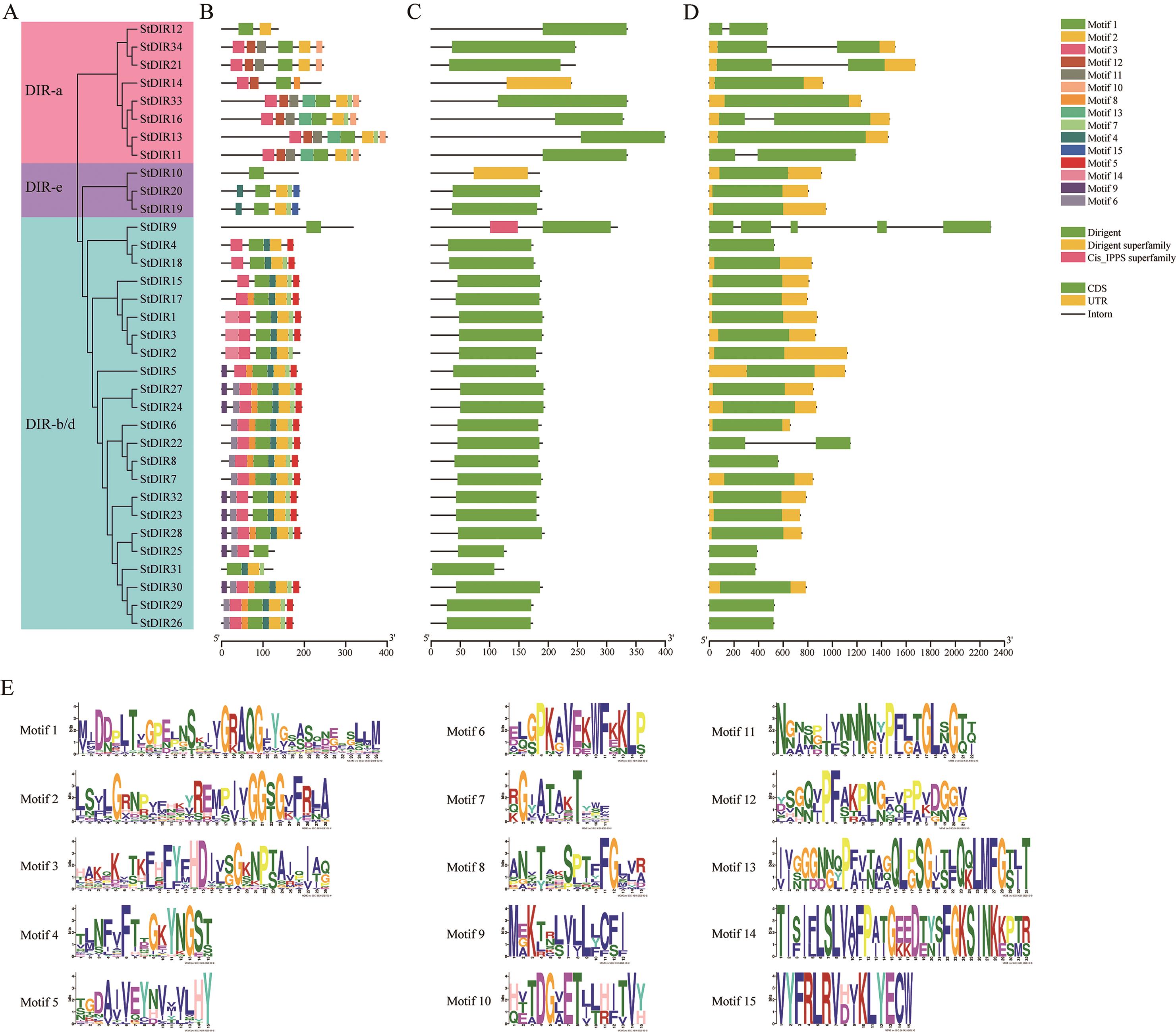
Fig. 3 Analysis of conserved motifs, domains and gene structure of DIRs in potatoA: Phylogenetic tree of StDIRs. B: Distribution of conserved motifs of StDIR proteins. C: The location of the conserved DIR domain. D: Structure of StDIR gene. E: Sequence information for each motif

Fig. 4 Collinearity analysis of DIR genes in potatoThe blue refers to links tandem duplication gene pairs, red refers to links segmental duplication gene pairs. The chromosomes, distributions of unknown bases, distributions of GC content, and the genome-wide gene density are demonstrated by rings from the inside to the outside, respectively
| 1 | Davin LB, Wang HB, Crowell AL, et al. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center [J]. Science, 1997, 275(5298): 362-366. |
| 2 | Burlat V, Kwon M, Davin LB, et al. Dirigent proteins and dirigent sites in lignifying tissues [J]. Phytochemistry, 2001, 57(6): 883-897. |
| 3 | Ralph S, Park JY, Bohlmann J, et al. Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.) [J]. Plant Mol Biol, 2006, 60(1): 21-40. |
| 4 | Davin LB, Lewis NG. Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis [J]. Plant Physiol, 2000, 123(2): 453-462. |
| 5 | Ralph SG, Jancsik S, Bohlmann J. Dirigent proteins in conifer defense Ⅱ extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.) [J]. Phytochemistry, 2007, 68(14): 1975-1991. |
| 6 | Davin LB, Jourdes M, Patten AM, et al. Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis [J]. Nat Prod Rep, 2008, 25(6):1015-1090. |
| 7 | Wu RH, Wang LL, Wang Z, et al. Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica [J]. Prog Nat Sci, 2009, 19(3): 347-352. |
| 8 | Li Q, Chen JF, Xiao Y, et al. The dirigent multigene family in Isatis indigotica: gene discovery and differential transcript abundance [J]. BMC Genomics, 2014, 15(1): 388. |
| 9 | Halls SC, Davin LB, Kramer DM, et al. Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein [J]. Biochemistry, 2004, 43(9): 2587-2595. |
| 10 | Pellegrini N, Valtueña S, Ardigò D, et al. Intake of the plant lignans matairesinol, secoisolariciresinol, pinoresinol, and lariciresinol in relation to vascular inflammation and endothelial dysfunction in middle age-elderly men and post-menopausal women living in Northern Italy [J]. Nutr Metab Cardiovasc Dis, 2010, 20(1): 64-71. |
| 11 | Ascacio-Valdés J, Buenrostro-Figueroa JJ, Aguilera-Carbó A, et al. Ellagitannins: Biosynthesis, biodegradation and biological properties [J]. J Med Plants Res, 2011, 5: 4696-4703. |
| 12 | Zhan ZJ, Ying YM, Ma LF, et al. Natural disesquiterpenoids [J]. Nat Prod Rep, 2011, 28(3): 594-629. |
| 13 | Rivière C, Pawlus AD, Mérillon JM. Natural stilbenoids: distribution in the plant Kingdom and chemotaxonomic interest in Vitaceae [J]. Nat Prod Rep, 2012, 29(11): 1317-1333. |
| 14 | Satake H, Koyama T, Bahabadi SE, et al. Essences in metabolic engineering of lignan biosynthesis [J]. Metabolites, 2015, 5(2): 270-290. |
| 15 | Paniagua C, Bilkova A, Jackson P, et al. Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure [J]. J Exp Bot, 2017, 68(13): 3287-3301. |
| 16 | Umezawa T, Davin LB, Yamamoto E, et al. Lignan biosynthesis in Forsythia species [J]. J Chem Soc, Chem Commun, 1990(20): 1405. |
| 17 | Cheng X, Su XQ, Muhammad A, et al. Molecular characterization, evolution, and expression profiling of the Dirigent (DIR) family genes in Chinese white pear (Pyrus bretschneideri) [J]. Front Genet, 2018, 9: 136. |
| 18 | Khan A, Li RJ, Sun JT, et al. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses [J]. Sci Rep, 2018, 8(1): 5500. |
| 19 | Liu ZW, Wang XF, Sun ZW, et al. Evolution, expression and functional analysis of cultivated allotetraploid cotton DIR genes [J]. BMC Plant Biol, 2021, 21(1): 89. |
| 20 | Hosmani PS, Kamiya T, Danku J, et al. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root [J]. Proc Natl Acad Sci USA, 2013, 110(35): 14498-14503. |
| 21 | Corbin C, Drouet S, Markulin L, et al. A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: from gene identification and evolution to differential regulation [J]. Plant Mol Biol, 2018, 97(1/2): 73-101. |
| 22 | Chou KC, Shen HB. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization [J]. PLoS One, 2010, 5(6): e11335. |
| 23 | Gupta R, Jung E, Brunak S. Prediction of N-glycosylation sites in human proteins [J]. 2004, 46: 203-206. |
| 24 | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| 25 | Zhang KJ, Xing WJ, Sheng SA, et al. Genome-wide identification and expression analysis of eggplant DIR gene family in response to biotic and abiotic stresses [J]. Horticulturae, 2022, 8(8): 732. |
| 26 | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching [J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| 27 | Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server [J]. Bioinformatics, 2015, 31(8): 1296-1297. |
| 28 | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| 29 | Thamil Arasan SK, Park JI, Ahmed NU, et al. Characterization and expression analysis of dirigent family genes related to stresses in Brassica [J]. Plant Physiol Biochem, 2013, 67: 144-153. |
| 30 | Liao YR, Liu SB, Jiang YY, et al. Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa) [J]. Genes Genom, 2017, 39(1): 47-62. |
| 31 | Song M, Peng XY. Genome-wide identification and characterization of DIR genes in Medicago truncatula [J]. Biochem Genet, 2019, 57(4): 487-506. |
| 32 | Li LL, Sun WB, Zhou PJ, et al. Genome-wide characterization of dirigent proteins in Populus: gene expression variation and expression pattern in response to Marssonina Brunnea and phytohormones [J]. Forests, 2021, 12(4): 507. |
| 33 | Ma XF, Xu WY, Liu T, et al. Functional characterization of soybean (Glycine max) DIRIGENT genes reveals an important role of GmDIR27 in the regulation of pod dehiscence [J]. Genomics, 2021, 113(1 Pt 2): 979-990. |
| 34 | Yadav V, Wang ZY, Yang XZ, et al. Comparative analysis, characterization and evolutionary study of dirigent gene family in Cucurbitaceae and expression of novel dirigent peptide against powdery mildew stress [J]. Genes, 2021, 12(3): 326. |
| 35 | 张洪伟. 马铃薯晚疫病抗性相关基因StDIR1和StDIR2的分离和表达 [D]. 保定: 河北大学, 2012. |
| Zhang HW. Isolation and expression of StDIR1 and StDIR2 genes related to potato late blight resistance [D]. Baoding: Hebei University, 2012. | |
| 36 | Liu JG, Stipanovic RD, Bell AA, et al. Stereoselective coupling of hemigossypol to form (+)-gossypol in moco cotton is mediated by a dirigent protein [J]. Phytochemistry, 2008, 69(18): 3038-3042. |
| 37 | Effenberger I, Zhang B, Li L, et al. Dirigent proteins from cotton (Gossypium sp.) for the atropselective synthesis of gossypol [J]. Angew Chem Int Ed, 2015, 54(49): 14660-14663. |
| 38 | Sahani P, Ujinwal M, Singh S, et al. Genome wide in silico characterization of dirigent protein family in flax (Linum usitatissmum l.) [J]. Plant Archives, 2018, 18: 61-68. |
| 39 | Ma RF, Huang B, Chen JL, et al. Genome-wide identification and expression analysis of dirigent-jacalin genes from plant chimeric lectins in Moso bamboo (Phyllostachys edulis) [J]. PLoS One, 2021, 16(3): e0248318. |
| 40 | Kim MK, Jeon JH, Fujita M, et al. The western red cedar (Thuja plicata) 8-8' DIRIGENT family displays diverse expression patterns and conserved monolignol coupling specificity [J]. Plant Mol Biol, 2002, 49(2): 199-214. |
| 41 | 马金洋, 杨瑾冬, 李卿, 等. 丹参Dirigent基因家族的发现与生物信息学分析 [J]. 基因组学与应用生物学, 2017, 36(4): 1594-1610. |
| Ma JY, Yang JD, Li Q, et al. Discovery and bioinformatics analysis of dirigent multigene family in Salvia miltiorrhiza [J]. Genom Appl Biol, 2017, 36(4): 1594-1610. | |
| 42 | 刘正文. 棉花纤维发育相关基因挖掘及DIR和GH9基因家族研究 [D]. 保定: 河北农业大学, 2021. |
| Liu ZW. Mining genes related to cotton fiber development and study on DIR and GH9 gene family [D]. Baoding: Hebei Agricultural University, 2021. | |
| 43 | 穰中文, 周清明. 水稻dirigent基因家族生物信息学分析 [J]. 湖南农业大学学报: 自然科学版, 2013, 39(2): 111-120. |
| Rang ZW, Zhou QM. Bioinformatic analysis of the dirigent gene family in rice [J]. J Hunan Agric Univ Nat Sci, 2013, 39(2): 111-120. | |
| 44 | Zhu LF, Zhang XL, Tu LL, et al. Isolation and characterization of two novel dirigent-like genes highly induced in cotton (Gossypium barbadense and G. hirsutum) after infection by Verticillium dahliae [J]. J Plant Pathol, 2007, 89: 41-45. |
| 45 | 关瑞攀. Dirigent基因参与三七-茄腐镰刀菌互作的分子机理研究 [D]. 昆明: 昆明理工大学, 2018. |
| Guan RP. Molecular mechanism of Dirigent genes involved in the interaction of Panax notoginseng-Fusarium solani [D]. Kunming: Kunming University of Science and Technology, 2018. | |
| 46 | Guo JL, Xu LP, Fang JP, et al. A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses [J]. Plant Cell Rep, 2012, 31(10): 1801-1812. |
| 47 | Shi YQ, Shen YR, Ahmad B, et al. Genome-wide identification and expression analysis of dirigent gene family in strawberry (Fragaria vesca) and functional characterization of FvDIR13 [J]. Sci Hortic, 2022, 297: 110913. |
| [1] | JIA Zi-jian, WANG Bao-qiang, CHEN Li-fei, WANG Yi-zhen, WEI Xiao-hong, ZHAO Ying. Expression Patterns of CHX Gene Family in Quinoa in Response to NO under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 163-174. |
| [2] | YAN Wei, CHEN Hui-ting, YE Qing, LIU Guang-chao, LIU Xin, HOU Li-xia. Identification of the Grape HCT Gene Family and Their Responses to Low-temperature Stress [J]. Biotechnology Bulletin, 2025, 41(2): 175-186. |
| [3] | KUANG Jian-hua, CHENG Zhi-peng, ZHAO Yong-jing, YANG Jie, CHEN Run-qiao, CHEN Long-qing, HU Hui-zhen. Expression Analysis of the GH3 Gene Family in Nelumbo nucifera underHormonal and Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(2): 221-233. |
| [4] | HUANG Ying, YU Wen-jing, LIU Xue-feng, DIAO Gui-ping. Bioinformatics and Expression Pattern Analysis of Glutathione S-transferase in Populus davidiana × P. bolleana [J]. Biotechnology Bulletin, 2025, 41(2): 248-256. |
| [5] | YANG Yong, YUAN Guo-mei, KANG Xiao-xiao, LIU Ya-ming, WANG Dong-sheng, ZHANG Hai-e. Identification and Expression Analysis of Members of the SWEET Gene Family in Chinese Chestnut [J]. Biotechnology Bulletin, 2025, 41(2): 257-269. |
| [6] | YANG Yong, CAO Rui, KANG Xiao-xiao, LIU Jing, WANG Xuan, ZHANG Hai-e. Identification and Expression Analysis of 13 Gene Families in the Chestnut Flavonoid Synthesis Pathway [J]. Biotechnology Bulletin, 2025, 41(2): 270-283. |
| [7] | LI Ming, LIU Xiang-yu, WANG Yi-na, HE Si-mei, SHA Ben-cai. Cloning and Functional Characterization of 6-OMT Gene Related to Isocorydine Biosynthesis in Dactylicapnos scandens [J]. Biotechnology Bulletin, 2025, 41(2): 309-320. |
| [8] | GE Shi-jie, LIU Yi-de, ZHANG Hua-dong, NING Qiang, ZHU Zhan-wang, WANG Shu-ping, LIU Yi-ke. Identification and Expression Analysis of Protein Disulfide Isomerase Gene Family in Wheat [J]. Biotechnology Bulletin, 2025, 41(2): 85-96. |
| [9] | YIN Yuan, CHENG Shuang, LIU Ding-hao, DENG Xiao-xia, LI Kai-yue, WANG Jing-hong, LIN Ji-xiang. Research Progress in Exogenous Hydrogen Peroxide(H2O2)Affecting Plant Growth and Physiological Metabolism under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(1): 1-13. |
| [10] | DU Pin-ting, WU Guo-jiang, WANG Zhen-guo, LI Yan, ZHOU Wei, ZHOU Ya-xing. Identification and Expression Analysis of CPP Gene Family in Sorghum [J]. Biotechnology Bulletin, 2025, 41(1): 132-142. |
| [11] | WU Zhi-jian, LIU Guang-yang, LIN Zhi-hao, SHENG Bin, CHEN Ge, XU Xiao-min, WANG Jun-wei, XU Dong-hui. Research Progress of Nano-regulation of Vegetable Seed Germination and Its Mechanism [J]. Biotechnology Bulletin, 2025, 41(1): 14-24. |
| [12] | LI Yu-xin, LI Miao, DU Xiao-fen, HAN Kang-ni, LIAN Shi-chao, WANG Jun. Identification and Expression Analysis of SiSAP Gene Family in Foxtail Millet(Setaria italica) [J]. Biotechnology Bulletin, 2025, 41(1): 143-156. |
| [13] | WANG Zi-ao, TIAN Rui, CUI Yong-mei, BAI Yi-xiong, YAO Xiao-hua, AN Li-kun, WU Kun-lun. Bioinformatics and Expression Pattern Analysis of HvnJAZ4 Gene in Hulless Barley [J]. Biotechnology Bulletin, 2025, 41(1): 173-185. |
| [14] | KONG Qing-yang, ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le. Identification and Expression Analysis of GRAS Transcription Factor Family in Rosa persica [J]. Biotechnology Bulletin, 2025, 41(1): 210-220. |
| [15] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||