








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (8): 234-241.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0059
LI Kai-jie1( ), WU Yao1, LI Dan-dan1,2(
), WU Yao1, LI Dan-dan1,2( )
)
Received:2025-01-14
Online:2025-08-26
Published:2025-08-14
Contact:
LI Dan-dan
E-mail:982641736@qq.com;ddli3@gzu.edu.cn
LI Kai-jie, WU Yao, LI Dan-dan. Cloning of Gene CtbHLH128 in Safflower and Response Function Regulating Drought Stress[J]. Biotechnology Bulletin, 2025, 41(8): 234-241.
名称 Primer name | 核苷酸序列 Nucleotide sequence (5′‒3′) | 功能/基因名 Function/Gene name |
|---|---|---|
| PF1 | ATGTATCCGAACTCCAATTCCTCT | Sequence clone (CtbHLH128) |
| PR1 | TCATCTTGGTTTGCATCCACACGAA | Sequence clone (CtbHLH128) |
| PF2 | atttggagaggacagggtaccATGTATCCGAACTCCAATTCCTCT | Recombination (CtbHLH128) |
| PR2 | cttgctcaccatggtactagtTCAAACCTTAGACCGTCCACCACCA | Recombination (CtbHLH128) |
| PF3 | GGTAATATCCGGAAACCTCCTCGGA | PCR identification (CtbHLH128) |
| PR3 | GCGGGCGAGACCTAATCCTCCGCCT | PCR identification (CtbHLH128) |
| PF4 | TAGATTCTTCTCACCTCCGC | semi-quantitative PCR (CtbHLH128) |
| PR4 | GCTTGGCGCGAATTTTACAAG | semi-quantitative PCR (CtbHLH128) |
| PF5 | GAACGGGAAATTGTCCGTG | semi-quantitative PCR (Atactin) |
| PR5 | TGAACAATCGATGGACCTGAC | semi-quantitative PCR (Atactin) |
Table 1 Information of primers in experiement
名称 Primer name | 核苷酸序列 Nucleotide sequence (5′‒3′) | 功能/基因名 Function/Gene name |
|---|---|---|
| PF1 | ATGTATCCGAACTCCAATTCCTCT | Sequence clone (CtbHLH128) |
| PR1 | TCATCTTGGTTTGCATCCACACGAA | Sequence clone (CtbHLH128) |
| PF2 | atttggagaggacagggtaccATGTATCCGAACTCCAATTCCTCT | Recombination (CtbHLH128) |
| PR2 | cttgctcaccatggtactagtTCAAACCTTAGACCGTCCACCACCA | Recombination (CtbHLH128) |
| PF3 | GGTAATATCCGGAAACCTCCTCGGA | PCR identification (CtbHLH128) |
| PR3 | GCGGGCGAGACCTAATCCTCCGCCT | PCR identification (CtbHLH128) |
| PF4 | TAGATTCTTCTCACCTCCGC | semi-quantitative PCR (CtbHLH128) |
| PR4 | GCTTGGCGCGAATTTTACAAG | semi-quantitative PCR (CtbHLH128) |
| PF5 | GAACGGGAAATTGTCCGTG | semi-quantitative PCR (Atactin) |
| PR5 | TGAACAATCGATGGACCTGAC | semi-quantitative PCR (Atactin) |

Fig. 2 Phylogenetic analysis of CtbHLH128A: Phylogenetic tree. B: Multiple-sequence alignment and analysis of the bHLH motif of amino acid sequences between CtbHLH128 and the closely related species. Amt: Amaranthus tricolor; Brd: Brachypodium distachyon; Brn: Brassica napus; Caa: Capsicum annuum; Cis: Citrus sinensis; Cus: Cucumis sativus; Eug: Eucalyptus grandis; Glm: Glycine max; Goh: Gossypium hirsutum; Hea: Helianthus annuus; Hov: Hordeum vulgare; Jur: Juglans regia; Kln: Klebsormidium nitens; Las: Lactuca sativa; Mae: Manihot esculenta; Met: Medicago truncatula; Nen: Nelumbo nucifera; Nit: Nicotiana tabacum; Ors: Oryza sativa; Php: Physcomitrium patens; Pot: Populus trichocarpa; Ric: Ricinus communis; Sei: Setaria italica; Sly: Solanum lycopersicum; Stu: Solanum tuberosum; Zma: Zea mays; Sbi: Sorghum bicolor; Ct: Carthamus tinctorius
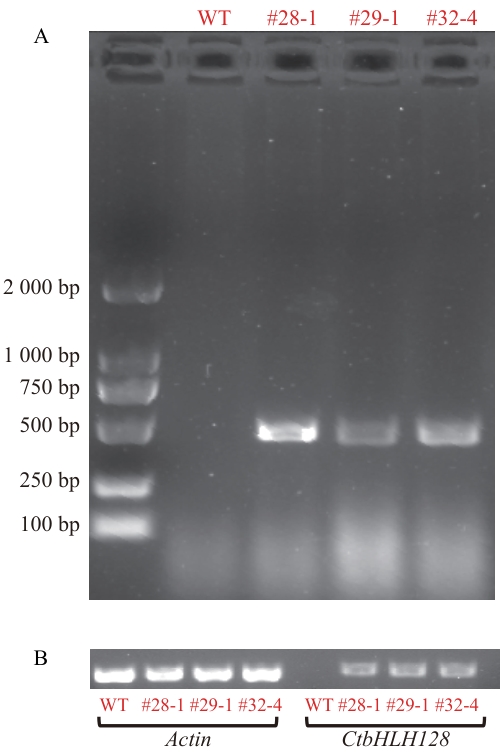
Fig. 3 Identification of positive transfected A. thalianaA: PCR identification results of the positive seedlings genomic DNA. B: Results of the semi-quantitative PCR between transgenic positive seedlings and control seedlings. #28-1, #29-1 and #32-4 are homozygous T3 positive lines transferred CtbHLH128 to Arabidopsis thaliana, and WT is wild-type. The same below

Fig. 4 Morphological diagram of overexpressed and wild-type Arabidopsis under drought stress during seedling stageOEbHLH#26, OEbHLH#7, OEbHLH#23, OEbHLH#3 and OEbHLH#36 indicate the different positive T2 lines. WT (col-gl) refers to wild type. The observation was conducted at 14 d after drought stress
| [1] | 吕培霖, 李成义, 彭文化, 等. 红花籽油抗氧化活性实验研究 [J]. 西北国防医学杂志, 2017, 38(7): 439-441. |
| Lv PL, Li CY, Peng WH, et al. Antioxidant activity of safflower seed oil [J]. Med J Natl Defending Forces Northwest China, 2017, 38(7): 439-441. | |
| [2] | 白启荣, 郭姣洁, 吴娇. 红花的化学成分及药理作用研究进展 [J]. 新乡医学院学报, 2024, 41(1): 88-94, 100. |
| Bai QR, Guo JJ, Wu J. Research progress on chemical constituents and pharmacological effects of safflower [J]. J Xinxiang Med Univ, 2024, 41(1): 88-94, 100. | |
| [3] | 吕培霖, 李成义, 王俊丽. 红花籽油的研究进展 [J]. 中国现代中药, 2016, 18(3): 387-389. |
| Lyu PL, Li CY, Wang JL. Review of safflower seed oil [J]. Mod Chin Med, 2016, 18(3): 387-389. | |
| [4] | 任超翔, 吴沂芸, 唐小慧, 等. 红花的起源与产地变迁 [J]. 中国中药杂志, 2017, 42(11): 2219-2222. |
| Ren CX, Wu YY, Tang XH, et al. Safflower's origin and changes of producing areas [J]. China J Chin Mater Med, 2017, 42(11): 2219-2222. | |
| [5] | 郭美丽, 张汉明, 张美玉. 红花本草考证 [J]. 中药材, 1996, 19(4): 202-203. |
| Guo ML, Zhang HM, Zhang MY. Herbological study of Honghua [J]. J Chin Med Mater, 1996, 19(4): 202-203. | |
| [6] | Gong ZZ, Xiong LM, Shi HZ, et al. Plant abiotic stress response and nutrient use efficiency [J]. Sci China Life Sci, 2020, 63(5): 635-674. |
| [7] | Xie YP, Chen PX, Yan Y, et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple [J]. New Phytol, 2018, 218(1): 201-218. |
| [8] | Samira R, Li BH, Kliebenstein D, et al. The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis [J]. Plant Mol Biol, 2018, 97(4/5): 297-309. |
| [9] | Govind G, Harshavardhan VT, Patricia JK, et al. Identification and functional validation of a unique set of drought induced genes preferentially expressed in response to gradual water stress in peanut [J]. Mol Genet Genomics, 2009, 281(6): 591-605. |
| [10] | Jin JP, Zhang H, Kong L, et al. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors [J]. Nucleic Acids Res, 2014, 42(Database issue): D1182-D1187. |
| [11] | Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity [J]. Mol Biol Evol, 2003, 20(5): 735-747. |
| [12] | Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors [J]. Plant J, 2011, 66(1): 94-116. |
| [13] | Bailey PC, Martin C, Toledo-Ortiz G, et al. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana [J]. Plant Cell, 2003, 15(11): 2497-2502. |
| [14] | Anuj Kumer Das. bHLH转录因子ABP7-Like在玉米叶片和籽粒发育中的功能鉴定和分子机制分析 [D]. 北京: 中国农业科学院, 2019. |
| Anuj KD. Functional characterization and molecular mechanism analysis of ABP7-like, a bHLH transcription factor, in maize leaf and kernel development [D]. Beijing: Chinese Academy of Agricultural Sciences, 2019. | |
| [15] | 汪德州, 莫小婷, 张霞, 等. 玉米转录因子ZmbHLH4基因的克隆及功能分析 [J]. 中国农业科技导报, 2018, 20(12): 16-25. |
| Wang DZ, Mo XT, Zhang X, et al. Cloning and functional analysis of transcription factors gene ZmbHLH4 from Zea mays [J]. J Agric Sci Technol, 2018, 20(12): 16-25. | |
| [16] | 刘玮. 花生AhbHLH1参与调控FAD2基因在种子中表达的研究 [D]. 济南: 山东大学, 2016. |
| Liu W. The study on AhbHLH1 involved in regulating the FAD2 gene expression in peanut (Arachis hypogaea L.) seed [D]. Jinan: Shandong University, 2016. | |
| [17] | 许红梅, 张立军, 刘淳. 农杆菌蘸花法侵染拟南芥的研究 [J]. 北方园艺, 2010(14): 143-146. |
| Xu HM, Zhang LJ, Liu C. Study of infection Arabidopsis thaliana by Agrobacterium dipping flower method [J]. North Hortic, 2010(14): 143-146. | |
| [18] | Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family [J]. Plant Cell, 2003, 15(8): 1749-1770. |
| [19] | Li XX, Duan XP, Jiang HX, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis [J]. Plant Physiol, 2006, 141(4): 1167-1184. |
| [20] | Zhang TT, Lv W, Zhang HS, et al. Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize [J]. BMC Plant Biol, 2018, 18(1): 235. |
| [21] | 李彩霞, 李艺, 穆宏秀, 等. 中国南瓜bHLH转录因子家族的鉴定与生物信息学分析 [J]. 生物技术通报, 2025, 41(1): 186-197. |
| Li CX, Li Y, Mu HX, et al. Identification and bioinformatics analysis of the bHLH transcription factor family in Cucurbita moschata duch [J]. Biotechnol Bull, 2025, 41(1): 186-197. | |
| [22] | 谢荟清, 苏文娟, 赵娜红, 等. 油茶bHLH基因家族鉴定与响应干旱胁迫的基因表达分析 [J]. 中南林业科技大学学报, 2025, 45(1): 168-180. |
| Xie HQ, Su WJ, Zhao NH, et al. Identification of bHLH gene family in Camellia oleifera and analysis of its expression under drought stress [J]. J Cent South Univ For Technol, 2025, 45(1): 168-180. | |
| [23] | Tan ZW, Lu DD, Yu YL, et al. Genome-wide identification and characterization of the bHLH gene family and its response to abiotic stresses in Carthamus tinctorius [J]. Plants, 2023, 12(21): 3764. |
| [24] | 刘福, 陈诚, 张凯旋, 等. 日本百脉根LjbHLH34基因克隆及耐旱功能鉴定 [J]. 草业学报, 2023, 32(1): 178-191. |
| Liu F, Chen C, Zhang KX, et al. Cloning and identification of drought tolerance function of the LjbHLH34 gene in Lotus japonicus [J]. Acta Prataculturae Sin, 2023, 32(1): 178-191. | |
| [25] | 张斌. 抗氧化途径下大豆GmbHLH130转录因子增强植物耐旱性研究 [J]. 西北农业学报, 2024, 33(9): 1746-1756. |
| Zhang B. Research of soybean GmbHLH130 transcription factor on plant drought tolerance under antioxidant pathway [J]. Acta Agric Boreali Occidentalis Sin, 2024, 33(9): 1746-1756. | |
| [26] | Takahashi Y, Ebisu Y, Kinoshita T, et al. bHLH transcription factors that facilitate K⁺ uptake during stomatal opening are repressed by abscisic acid through phosphorylation [J]. Sci Signal, 2013, 6(280): ra48. |
| [27] | 鄢梦雨, 韦晓薇, 曹婧, 等. 异子蓬SabHLH169基因的克隆及抗旱功能分析 [J]. 生物技术通报, 2023, 39(11): 328-339. |
| Yan MY, Wei XW, Cao J, et al. Cloning of basic helix-loop-helix(bHLH) transcription factor gene SabHLH169 in Suaeda aralocaspica and analysis of its resistances to drought stress [J]. Biotechnol Bull, 2023, 39(11): 328-339. | |
| [28] | Zhao PC, Li XX, Jia JT, et al. bHLH92 from sheepgrass acts as a negative regulator of anthocyanin/proanthocyandin accumulation and influences seed dormancy [J]. J Exp Bot, 2019, 70(1): 269-284. |
| [29] | 陶均, 李玲. 高等植物脱落酸生物合成的酶调控 [J]. 植物学通报, 2002, 37(6): 675-683. |
| Tao J, Li L. Abscisic acid biosynthesis regulated by some key enzymes in higher plants [J]. Chin Bull Bot, 2002, 37(6): 675-683. | |
| [30] | 周金鑫, 胡新文, 张海文, 等. ABA在生物胁迫应答中的调控作用 [J]. 农业生物技术学报, 2008, 16(1): 169-174. |
| Zhou JX, Hu XW, Zhang HW, et al. Regulatory role of ABA in plant response to biotic stresses [J]. J Agric Biotechnol, 2008, 16(1): 169-174. | |
| [31] | 郭秀林, 刘子会, 李运朝, 等. Ca2+/CaM对玉米干旱信息传递的介导作用研究 [J]. 作物学报, 2005, 31(8): 1001-1006. |
| Guo XL, Liu ZH, Li YC, et al. Function of Ca2+/CaM on transduction of stress signal [J]. Acta Agron Sin, 2005, 31(8): 1001-1006. |
| [1] | CHEN Qiang, YU Ying-fei, ZHANG Ying, ZHANG Chong. Regulatory Effect of Methyl Jasmonate on Postharvest Chilling Injury in Oriental Melon ‘Emerald’ [J]. Biotechnology Bulletin, 2025, 41(9): 1-10. |
| [2] | CHENG Xue, FU Ying, CHAI Xiao-jiao, WANG Hong-yan, DENG Xin. Identification of LHC Gene Family in Setaria italica and Expression Analysis under Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(8): 102-114. |
| [3] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [4] | LI Ya-qiong, GESANG La-mao, CHEN Qi-di, YANG Yu-huan, HE Hua-zhuan, ZHAO Yao-fei. Heterologous Overexpression of Sorghum SbSnRK2.1 Enhances the Resistance to Salt Stress in Arabidopsis [J]. Biotechnology Bulletin, 2025, 41(8): 115-123. |
| [5] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [6] | LI Xin-ni, LI Jun-yi, MA Xue-hua, HE Wei, LI Jia-li, YU Jia, CAO Xiao-ning, QIAO Zhi-jun, LIU Si-chen. Identification of the PMEI Gene Family of Pectin Methylesterase Inhibitor in Foxtail Millet and Analysis of Its Response to Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(7): 150-163. |
| [7] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [8] | LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 181-192. |
| [9] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [10] | ZHANG Ze, YANG Xiu-li, NING Dong-xian. Identification of 4CL Gene Family in Arachis hypogaea L. and Expression Analysis in Response to Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(7): 117-127. |
| [11] | CHENG Hui-juan, WANG Xin, SHI Xiao-tao, MA Dong-xu, GONG Da-chun, HU Jun-peng, XIE Zhi-wen. Effects of Transcription Factor CREA Knockout on the Morphology and the Secretion of β-glucosidase in Aspergillus niger [J]. Biotechnology Bulletin, 2025, 41(6): 344-354. |
| [12] | WANG Miao-miao, ZHAO Xiang-long, WANG Zhao-ming, LIU Zhi-peng, YAN Long-feng. Identification of TCP Gene Family in Medicago ruthenica and Their Expression Pattern Analysis under Drought Stress [J]. Biotechnology Bulletin, 2025, 41(6): 179-190. |
| [13] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [14] | LI Xiao-huan, CHEN Xiang-yu, TAO Qi-yu, ZHU Ling, TANG Ming, YAO Yin-an, WANG Li-jun. Effects of PtoMYB61 on Lignin Biosynthesis and Salt Tolerance in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(6): 284-296. |
| [15] | ZONG Jian-wei, DENG Hai-fang, CAI Yuan-yuan, CHANG Ya-wen, ZHU Ya-qi, YANG Yu-hua. Coupling Effect of AM Fungi on the Root Morphology and Leaf Structure of Xanthoceras sorbifolium Bunge under Drought Stress [J]. Biotechnology Bulletin, 2025, 41(6): 167-178. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||