








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 161-169.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0747
Previous Articles Next Articles
LYU Cheng-cong( ), HENG Meng, CHEN Si-qi, JIN Xue-hua(
), HENG Meng, CHEN Si-qi, JIN Xue-hua( )
)
Received:2025-07-11
Online:2026-01-26
Published:2026-02-04
Contact:
JIN Xue-hua
E-mail:lvccong@126.com;xhkim2021@163.com
LYU Cheng-cong, HENG Meng, CHEN Si-qi, JIN Xue-hua. Cloning and Functional Analysis of ZhGSTF Related to Anthocyanin Transport Zantedeschia hybrida[J]. Biotechnology Bulletin, 2026, 42(1): 161-169.

Fig. 1 Four varieties of Z. hybrida spatula (A) and four coloring stages of rose spatula (B)S1: The spatula is uncolored; S2: micro-coloring the spatula; S3: partially coloring the spathe; S4: completely coloring the spatula. The same below
引物 Primer | 引物序列 Primer sequence (5′‒3′) | 引物用途 Primer usage |
|---|---|---|
| ZhGSTF-F | TATGCCGTTACGTGTGCGAC | 基因克隆 |
| ZhGSTF-R | GGTTGTAGGACTGCCCCTCT | Gene clone |
| ZhGSTF-F-q | AGATCCGTATCAATCCCT | 基因表达 Gene expression |
| ZhGSTF-R-q | TAAGAGTATGAAAGTGCC | |
| ZhANS-F | GATCCCGTGGTTGACGATGT | |
| ZhANS-R | ATCGTGGACTTCACGTCTGG | |
| ZhANR-F | TGGAAGACATAGTCGCAGCC | |
| ZhANR-R | GACGAAGCTCGCTCACCTTA | |
| ZhDFR-F | CGACGAGATGAAAGGACCCA | |
| ZhDFR-R | TGTATCGAAGACGTTGGCGG | |
| ZhActin-F | GAAGGAGAAACTTGCGTATGTAGC | 内参基因 |
| ZhActin-R | GTCCATCTGGCAGCTCATAACT | Reference gene |
| ZhGSTF-Sac I-F | CTCTAGAAGGCCTCCATGGGGATCCGCAGAGGGGCAGTCCTACAA | 载体构建 |
| ZhGSTF-BamH I-R | GCCTCGAGACGCGTGAGCTCGGAGCCTTCTGCTTCTGCA | Vectors construction |
Table 1 Primers used in the study
引物 Primer | 引物序列 Primer sequence (5′‒3′) | 引物用途 Primer usage |
|---|---|---|
| ZhGSTF-F | TATGCCGTTACGTGTGCGAC | 基因克隆 |
| ZhGSTF-R | GGTTGTAGGACTGCCCCTCT | Gene clone |
| ZhGSTF-F-q | AGATCCGTATCAATCCCT | 基因表达 Gene expression |
| ZhGSTF-R-q | TAAGAGTATGAAAGTGCC | |
| ZhANS-F | GATCCCGTGGTTGACGATGT | |
| ZhANS-R | ATCGTGGACTTCACGTCTGG | |
| ZhANR-F | TGGAAGACATAGTCGCAGCC | |
| ZhANR-R | GACGAAGCTCGCTCACCTTA | |
| ZhDFR-F | CGACGAGATGAAAGGACCCA | |
| ZhDFR-R | TGTATCGAAGACGTTGGCGG | |
| ZhActin-F | GAAGGAGAAACTTGCGTATGTAGC | 内参基因 |
| ZhActin-R | GTCCATCTGGCAGCTCATAACT | Reference gene |
| ZhGSTF-Sac I-F | CTCTAGAAGGCCTCCATGGGGATCCGCAGAGGGGCAGTCCTACAA | 载体构建 |
| ZhGSTF-BamH I-R | GCCTCGAGACGCGTGAGCTCGGAGCCTTCTGCTTCTGCA | Vectors construction |
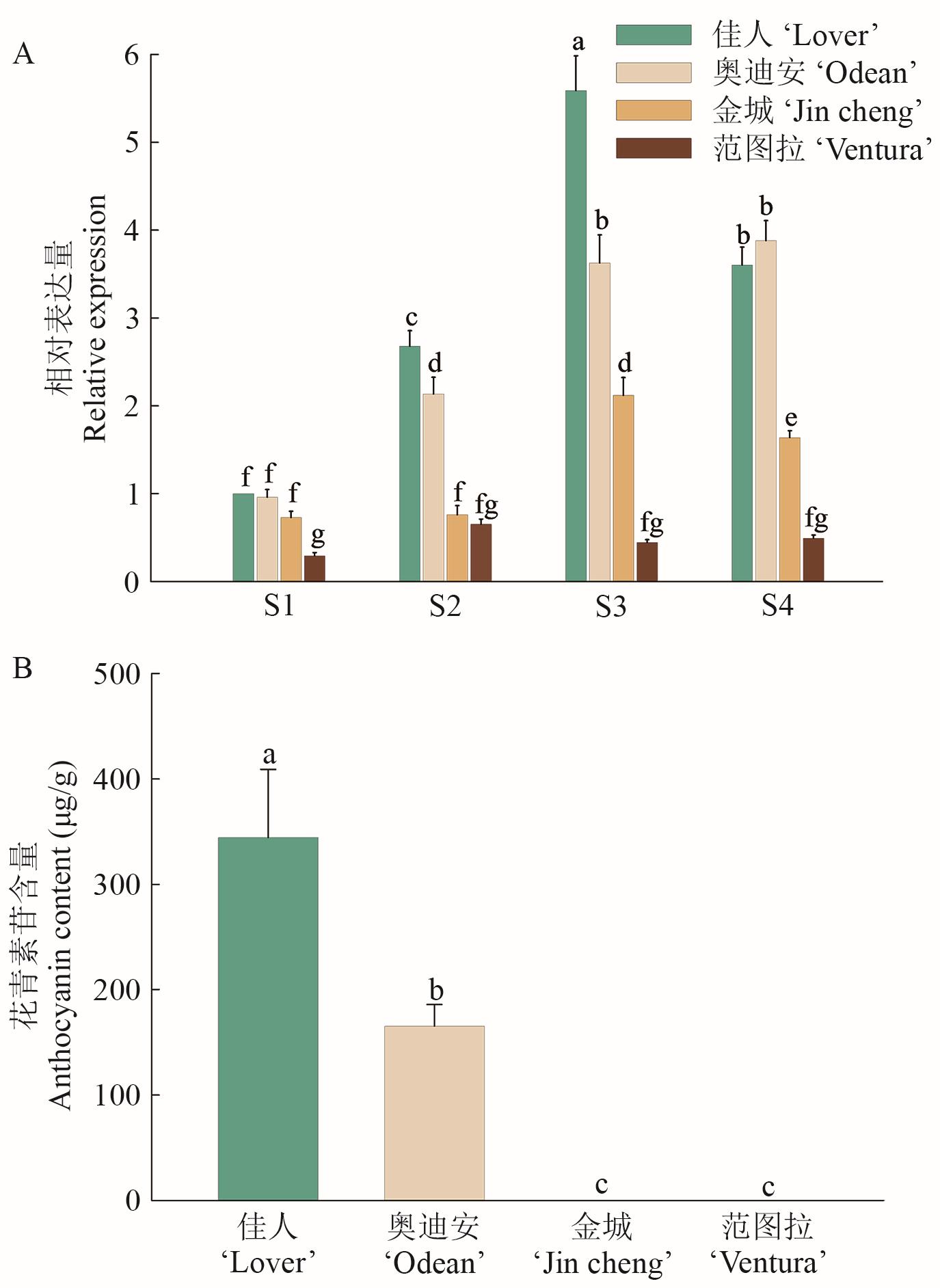
Fig. 4 Expressions of ZhGSTF in four coloring stages of four Z. hybrida varietiesA: The expressions of ZhGSTF in four coloring stages of four colored Z. hybrida varieties. B: Anthocyanin content in spatula of four different varieties of Z. hybrida at S4 stage. Different lowercase letters indicate significant differences among different treatments (P<0.05), the same below
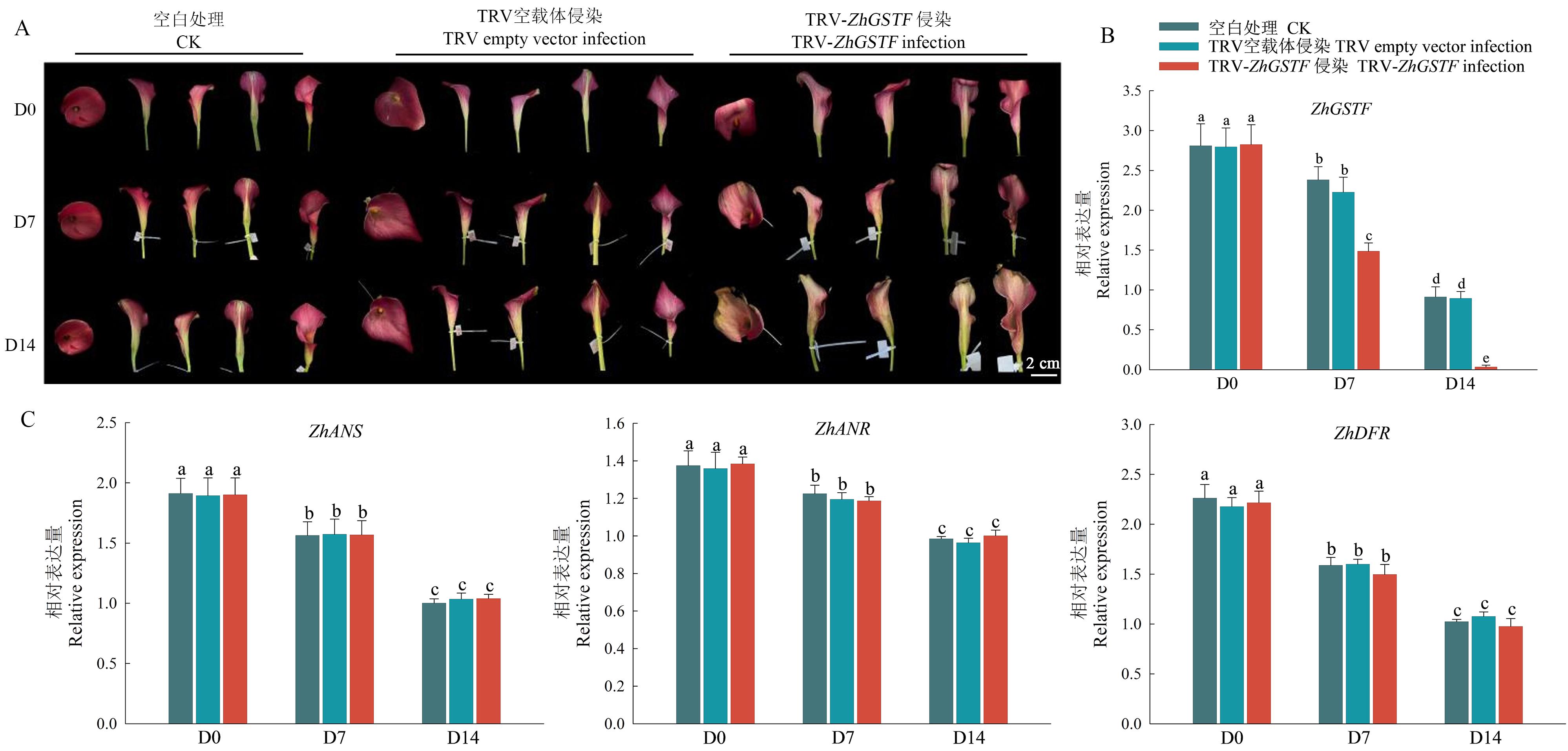
Fig. 6 Phenotype and related gene expression analysis of spathe after ZhGSTF silencingA: Phenotype of spathe after VIGS silencing ZhGSTF. B: Relative expression of ZhGSTF in spathe after VIGS silencing. C: Expressions of three key genes downstream of flavonoid synthesis pathway in spathe after ZhGSTF silencing. D0: 0 d of treatment; D7: 7th day of treatment; D4: 14th day of treatment
| [1] | Wang Y, Yang T, Wang D, et al. Chromosome level genome assembly of colored calla lily (Zantedeschia elliottiana) [J]. Sci Data, 2023, 10(1): 605. |
| [2] | 吴穷, 孙灿, 汤桂钧, 等. 上海地区彩色马蹄莲组织培养快繁及鲜切花周年生产技术 [J]. 上海农业科技, 2022(6): 91-93, 117. |
| Wu Q, Sun C, Tang GJ, et al. Tissue culture and rapid propagation of colored Calla lily and annual production techniques of fresh cut flowers in Shanghai [J]. Shanghai Agric Sci Technol, 2022(6): 91-93, 117. | |
| [3] | 李锐, 胡婷, 陈树溦, 等. 紫苏PfMYB80转录因子正向调控花青素的生物合成 [J]. 生物技术通报, 2025, 41(6): 243-255. |
| Li R, Hu T, Chen SW, et al. Positive regulation of anthocyanin biosynthesis by PfMYB80 transcription factor in Perilla frutescens [J]. Biotechnol Bull, 2025, 41(6): 243-255. | |
| [4] | Smeriglio A, Barreca D, Bellocco E, et al. Chemistry, pharmacology and health benefits of anthocyanins [J]. Phytother Res, 2016, 30(8): 1265-1286. |
| [5] | Cui YM, Fan JW, Lu CF, et al. ScGST3 and multiple R2R3-MYB transcription factors function in anthocyanin accumulation in Senecio cruentus [J]. Plant Sci, 2021, 313: 111094. |
| [6] | Lei T, Song Y, Jin XH, et al. Effects of pigment constituents and their distribution on spathe coloration of Zantedeschia hybrida [J]. HortScience, 2017, 52(12): 1840-1848. |
| [7] | Fang Y, Lei T, Wu YM, et al. Mechanism underlying color variation in calla lily spathes based on transcriptomic analysis [J]. J Amer Soc Hort Sci, 2021, 146(6): 387-398. |
| [8] | 赵婧, 郭茜, 李睿琦, 等. 大豆GmGST基因簇基因序列分析及诱导表达分析 [J]. 生物技术通报, 2025, 41(5): 129-140. |
| Zhao J, Guo Q, Li RQ, et al. Sequence analysis and induced expression analysis of GmGST gene cluster genes in soybean [J]. Biotechnol Bull, 2025, 41(5): 129-140. | |
| [9] | Manzoor MA, Ali Sabir I, Shah IH, et al. Flavonoids: a review on biosynthesis and transportation mechanism in plants [J]. Funct Integr Genomics, 2023, 23(3): 212. |
| [40] | Yang XM, Wang JJ, Wang DL, et al. Functional verification and bioinformatics analysis of cotton GhDMT3 gene [J]. Biotechnol Bull, 2019, 35(1): 11-16. |
| [41] | Tian LL, Liu LQ, Jiang YQ, et al. Tobacco rattle virus-induced VcANS gene silencing in blueberry fruit [J]. Gene, 2023, 852: 147054. |
| [42] | Zhang KJ, Wang XB, Chen XX, et al. Establishment of a homologous silencing system with intact-plant infiltration and minimized operation for studying gene function in herbaceous peonies [J]. Int J Mol Sci, 2024, 25(8): 4412. |
| [43] | 杨君, 孔羽, 刘群录, 等. 遮荫对‘花手鞠’绣球花色和花青素苷组成的影响 [J]. 园艺学报, 2023, 50(7): 1467-1481. |
| Yang J, Kong Y, Liu QL, et al. Effects of shading on the variation of sepal color and pigment composition in Hydrangea macrophylla ‘Hanatemari’ [J]. Acta Hortic Sin, 2023, 50(7): 1467-1481. | |
| [44] | 郑先哲, 赵兴隆, 刘成海, 等. 基于果实颜色特征的蓝靛果忍冬花青素含量预测 [J]. 农业工程学报, 2023, 39(2): 242-251. |
| Zheng XZ, Zhao XL, Liu CH, et al. Prediction of the anthocyanin content of Lonicera edulis based on fruit color characteristics [J]. Trans Chin Soc Agric Eng, 2023, 39(2): 242-251. | |
| [45] | 欧阳莎莎, 曹受金, 熊颖, 等. 不同樱花花瓣色值和花色素测定及抗氧化能力分析 [J]. 中南林业科技大学学报, 2024, 44(2): 166-173, 183. |
| Ouyang SS, Cao SJ, Xiong Y, et al. Measurement of color values and active substances of different flowering cherry petals and analysis of antioxidant capacity [J]. J Cent South Univ For Technol, 2024, 44(2): 166-173, 183. | |
| [46] | Han LL, Zhou L, Zou HZ, et al. PsGSTF3, an anthocyanin-related glutathione S-transferase gene, is essential for petal coloration in tree peony [J]. Int J Mol Sci, 2022, 23(3): 1423. |
| [47] | Huang CB, Zhao T, Li JH, et al. Glutathione transferase VvGSTU60 is essential for proanthocyanidin accumulation and cooperates synergistically with MATE in grapes [J]. Plant J, 2025, 121(2): e17197. |
| [10] | Hasan MS, Singh V, Islam S, et al. Genome-wide identification and expression profiling of glutathione S-transferase family under multiple abiotic and biotic stresses in Medicago truncatula L [J]. PLoS One, 2021, 16(2): e0247170. |
| [11] | Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases [J]. Genome Biol, 2002, 3(3): 3004.1. |
| [12] | Kitamura S, Shikazono N, Tanaka A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis [J]. Plant J, 2004, 37(1): 104-114. |
| [13] | Tasaki K, Yoshida M, Nakajima M, et al. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in Japanese gentian with the CRISPR/Cas9 system [J]. BMC Plant Biol, 2020, 20(1): 370. |
| [14] | Kitamura S, Akita Y, Ishizaka H, et al. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in Cyclamen [J]. J Plant Physiol, 2012, 169(6): 636-642. |
| [15] | Hu B, Zhao JT, Lai B, et al. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn [J]. Plant Cell Rep, 2016, 35(4): 831-843. |
| [16] | Pérez-Díaz R, Madrid-Espinoza J, Salinas-Cornejo J, et al. Differential roles for VviGST1, VviGST3, and VviGST4 in proanthocyanidin and anthocyanin transport in Vitis vinífera . [J]. Front Plant Sci, 2016, 7: 1166. |
| [17] | Luo HF, Dai C, Li YP, et al. Reduced anthocyanins in petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry [J]. J Exp Bot, 2018, 69(10): 2595-2608. |
| [18] | Jiang SH, Chen M, He NB, et al. MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple [J]. Hortic Res, 2019, 6: 40. |
| [19] | Zhao YW, Wang CK, Huang XY, et al. Genome-wide analysis of the glutathione S-transferase (GST) genes and functional identification of MdGSTU12 reveals the involvement in the regulation of anthocyanin accumulation in apple [J]. Genes, 2021, 12(11): 1733. |
| [20] | Zhao Y, Dong WQ, Zhu YC, et al. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach [J]. Plant Biotechnol J, 2020, 18(5): 1284-1295. |
| [21] | Lu ZH, Cao HH, Pan L, et al. Two loss-of-function alleles of the glutathione S-transferase (GST) gene cause anthocyanin deficiency in flower and fruit skin of peach (Prunus persica) [J]. Plant J, 2021, 107(5): 1320-1331. |
| [22] | Zhang T, Wu H, Sun YJ, et al. Identification of the GST gene family and functional analysis of RcGSTF2 related to anthocyanin in Rosa chinensis ‘old blush’ [J]. Plants, 2025, 14(6): 932. |
| [23] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [24] | 孙卫, 李崇晖, 王亮生, 等. 花青苷成分对瓜叶菊花色的影响 [J]. 园艺学报, 2009, 36(12): 1775-1782. |
| Sun W, Li CH, Wang LS, et al. Anthocyanins present in flowers of Senecio cruentus with different colors [J]. Acta Hortic Sin, 2009, 36(12): 1775-1782. | |
| [25] | 胡可, 韩科厅, 戴思兰. 环境因子调控植物花青素苷合成及呈色的机理 [J]. 植物学报, 2010, 45(3): 307-317. |
| Hu K, Han KT, Dai SL. Regulation of plant anthocyanin synthesis and pigmentation by environmental factors [J]. Chin Bull Bot, 2010, 45(3): 307-317. | |
| [26] | Saito K, Yonekura-Sakakibara K, Nakabayashi R, et al. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity [J]. Plant Physiol Biochem, 2013, 72: 21-34. |
| [27] | Gomez C, Conejero G, Torregrosa L, et al. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST [J]. Plant J, 2011, 67(6): 960-970. |
| [28] | 尹雨钦, 徐欢欢, 唐丽萍, 等. 不结球白菜GST基因家族的全基因组鉴定及花青素相关基因BcGSTF6的功能分析 [J]. 中国农业科学, 2024, 57(16): 3234-3249. |
| Yin YQ, Xu HH, Tang LP, et al. Genome-wide identification of GST gene family and functional analysis of the BcGSTF6 gene related to anthocyanin in pak choi [J]. Sci Agric Sin, 2024, 57(16): 3234-3249. | |
| [29] | Marrs KA, Alfenito MR, Lloyd AM, et al. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2 [J]. Nature, 1995, 375(6530): 397-400. |
| [30] | Yamazaki M, Shibata M, Nishiyama Y, et al. Differential gene expression profiles of red and green forms of Perilla frutescens leading to comprehensive identification of anthocyanin biosynthetic genes [J]. FEBS J, 2008, 275(13): 3494-3502. |
| [31] | Matsui K, Tomatsu T, Kinouchi S, et al. Identification of a gene encoding glutathione S-transferase that is related to anthocyanin accumulation in buckwheat (Fagopyrum esculentum) [J]. J Plant Physiol, 2018, 231: 291-296. |
| [32] | Lin YX, Zhang LX, Zhang JH, et al. Identification of anthocyanins-related glutathione S-transferase (GST) genes in the genome of cultivated strawberry (Fragaria × Ananassa) [J]. Int J Mol Sci, 2020, 21(22): 8708. |
| [33] | Xue L, Huang XR, Zhang ZH, et al. An anthocyanin-related glutathione S-transferase, MrGST1, plays an essential role in fruit coloration in Chinese bayberry (Morella rubra) [J]. Front Plant Sci, 2022, 13: 903333. |
| [34] | Qiu LK, Chen K, Pan J, et al. Genome-wide analysis of glutathione S-transferase genes in four Prunus species and the function of PmGSTF2, activated by PmMYBa1, in regulating anthocyanin accumulation in Prunus mume [J]. Int J Biol Macromol, 2024, 281: 136506. |
| [35] | Chen HX, Lei PH, Zhang HJ, et al. HmGST9, an anthocyanin-related glutathione S-transferase gene, is essential for sepals coloration in Hydrangea macrophylla [J]. J Plant Physiol, 2025, 307: 154466. |
| [36] | Zhang Z, Tian CP, Zhang Y, et al. Transcriptomic and metabolomic analysis provides insights into anthocyanin and procyanidin accumulation in pear [J]. BMC Plant Biol, 2020, 20(1): 129. |
| [37] | Grotewold E. The genetics and biochemistry of floral pigments [J]. Annu Rev Plant Biol, 2006, 57: 761-780. |
| [38] | Liu YH, Li YM, Liu Z, et al. Integrated transcriptomic and metabolomic analysis revealed altitude-related regulatory mechanisms on flavonoid accumulation in potato tubers [J]. Food Res Int, 2023, 170: 112997. |
| [39] | Rössner C, Lotz D, Becker A. VIGS goes viral: how VIGS transforms our understanding of plant science [J]. Annu Rev Plant Biol, 2022, 73: 703-728. |
| [40] | 杨笑敏, 王俊娟, 王德龙, 等. 棉花GhDMT3的功能验证及生物信息学分析 [J]. 生物技术通报, 2019, 35(1): 11-16. |
| [1] | YANG Juan, FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei. Cloning and Functional Analysis of AP2/ERF Transcription Factors RcERF4 and RcRAP2-12 in Rose [J]. Biotechnology Bulletin, 2026, 42(1): 150-160. |
| [2] | WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought [J]. Biotechnology Bulletin, 2026, 42(1): 198-207. |
| [3] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [4] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [5] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [6] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [7] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [8] | LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress [J]. Biotechnology Bulletin, 2025, 41(8): 220-233. |
| [9] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [10] | WANG Fang, QIAO Shuai, SONG Wei, CUI Peng-juan, LIAO An-zhong, TAN Wen-fang, YANG Song-tao. Genome-wide Identification of the IbNRT2 Gene Family and Its Expression in Sweet Potato [J]. Biotechnology Bulletin, 2025, 41(7): 193-204. |
| [11] | GUO Tao, AI Li-jiao, ZOU Shi-hui, ZHOU Ling, LI Xue-mei. Functional Study of CjRAV1 from Camellia japonica in Regulating Flowering Delay [J]. Biotechnology Bulletin, 2025, 41(6): 208-217. |
| [12] | ZHANG Yong, SONG Sheng-long, LI Yong-tai, ZHANG Xin-yu, LI Yan-jun. Cloning of GhSWEET9 in Upland Cotton and Functional Analysis of Resistance to Verticillium Wilt [J]. Biotechnology Bulletin, 2025, 41(6): 144-154. |
| [13] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [14] | LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 141-152. |
| [15] | ZHAO Jing, GUO Qian, LI Rui-qi, LEI Ying-yang, YUE Ai-qin, ZHAO Jin-zhong, YIN Cong-cong, DU Wei-jun, NIU Jing-ping. Sequence Analysis and Induced Expression Analysis of GmGST Gene Cluster Genes in Soybean [J]. Biotechnology Bulletin, 2025, 41(5): 129-140. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||