








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (5): 141-152.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0745
LI Zhi-qiang1( ), LUO Zheng-qian2, XU Lin-li2, ZHOU Guo-hui1, QU Si-yu1, LIU En-liang3(
), LUO Zheng-qian2, XU Lin-li2, ZHOU Guo-hui1, QU Si-yu1, LIU En-liang3( ), XU Dong-ting4(
), XU Dong-ting4( )
)
Received:2024-08-02
Online:2025-05-26
Published:2025-06-05
Contact:
LIU En-liang, XU Dong-ting
E-mail:adslizq@126.com;liuenliang_513@163.com;18636861516@163.com
LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress[J]. Biotechnology Bulletin, 2025, 41(5): 141-152.
引物名称 Primer name | 正向引物 Forward sequence (5'-3') | 反向引物 Reverse sequence (5'-3') |
|---|---|---|
| Gm_MYB114 | GGACCATACCTCCTTGGCAG | GGTGATGGTGGTGTGTCCAA |
| Gm_MYB137 | AGCTGCCAGGAAGAACAGAC | ACGCAACGTGTCTCTGAACT |
| Gm_MYB169 | TCCTCTGATGGAAGCTGCAC | TGGCTAAACCACTCACCAGC |
| Gm_MYB19 | ATCGGTGGGCCAAAATAGCA | TTAGCCTGCTTCACCACTCG |
| Gm_MYB208 | CCCAGTGACACACAGTCCTC | TACTGGTGCATGCCAAGGAG |
| Gm_MYB214 | TGGTTCAACAGGAAGTGCGT | GCTTGAGGCCAGGATGTAGG |
| Gm_MYB249 | GGCATGTGATGATGGCAAGG | GTGACACAAGGGTGGCTGTA |
| Gm_MYB51 | GGCAAAGGACATGGATCGGA | AACCAACCTCCGTAACGCTT |
| Gm_MYB76 | GCCAGGCAGAACAGACAATG | GGCATAGGAGTTGCCTGGTT |
| Gm_MYB89 | GATGGTCTACCATTGCGGCT | GTGTGGTGTGCCAATGGTTC |
| GmActin | CGGTGGTTCTATCTTGGCATC | GTCTTTCGCTTCAATAACCCTA |
Table 1 Primer information used
引物名称 Primer name | 正向引物 Forward sequence (5'-3') | 反向引物 Reverse sequence (5'-3') |
|---|---|---|
| Gm_MYB114 | GGACCATACCTCCTTGGCAG | GGTGATGGTGGTGTGTCCAA |
| Gm_MYB137 | AGCTGCCAGGAAGAACAGAC | ACGCAACGTGTCTCTGAACT |
| Gm_MYB169 | TCCTCTGATGGAAGCTGCAC | TGGCTAAACCACTCACCAGC |
| Gm_MYB19 | ATCGGTGGGCCAAAATAGCA | TTAGCCTGCTTCACCACTCG |
| Gm_MYB208 | CCCAGTGACACACAGTCCTC | TACTGGTGCATGCCAAGGAG |
| Gm_MYB214 | TGGTTCAACAGGAAGTGCGT | GCTTGAGGCCAGGATGTAGG |
| Gm_MYB249 | GGCATGTGATGATGGCAAGG | GTGACACAAGGGTGGCTGTA |
| Gm_MYB51 | GGCAAAGGACATGGATCGGA | AACCAACCTCCGTAACGCTT |
| Gm_MYB76 | GCCAGGCAGAACAGACAATG | GGCATAGGAGTTGCCTGGTT |
| Gm_MYB89 | GATGGTCTACCATTGCGGCT | GTGTGGTGTGCCAATGGTTC |
| GmActin | CGGTGGTTCTATCTTGGCATC | GTCTTTCGCTTCAATAACCCTA |
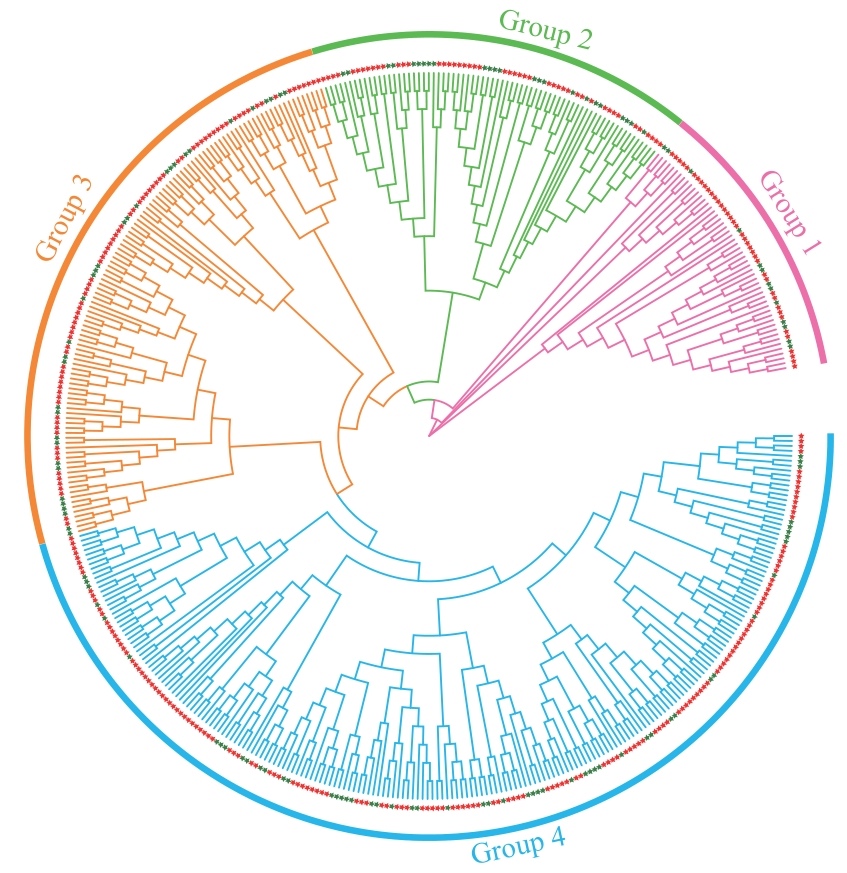
Fig. 2 Phylogenetic tree of soybean and Arabidopsis thalianaR2R3-MYB genesDifferent colors indicate different groups. The red star indicates the soybean R2R3-MYB gene, and the green star indicates the Arabidopsis thalianaR2R3-MYB gene
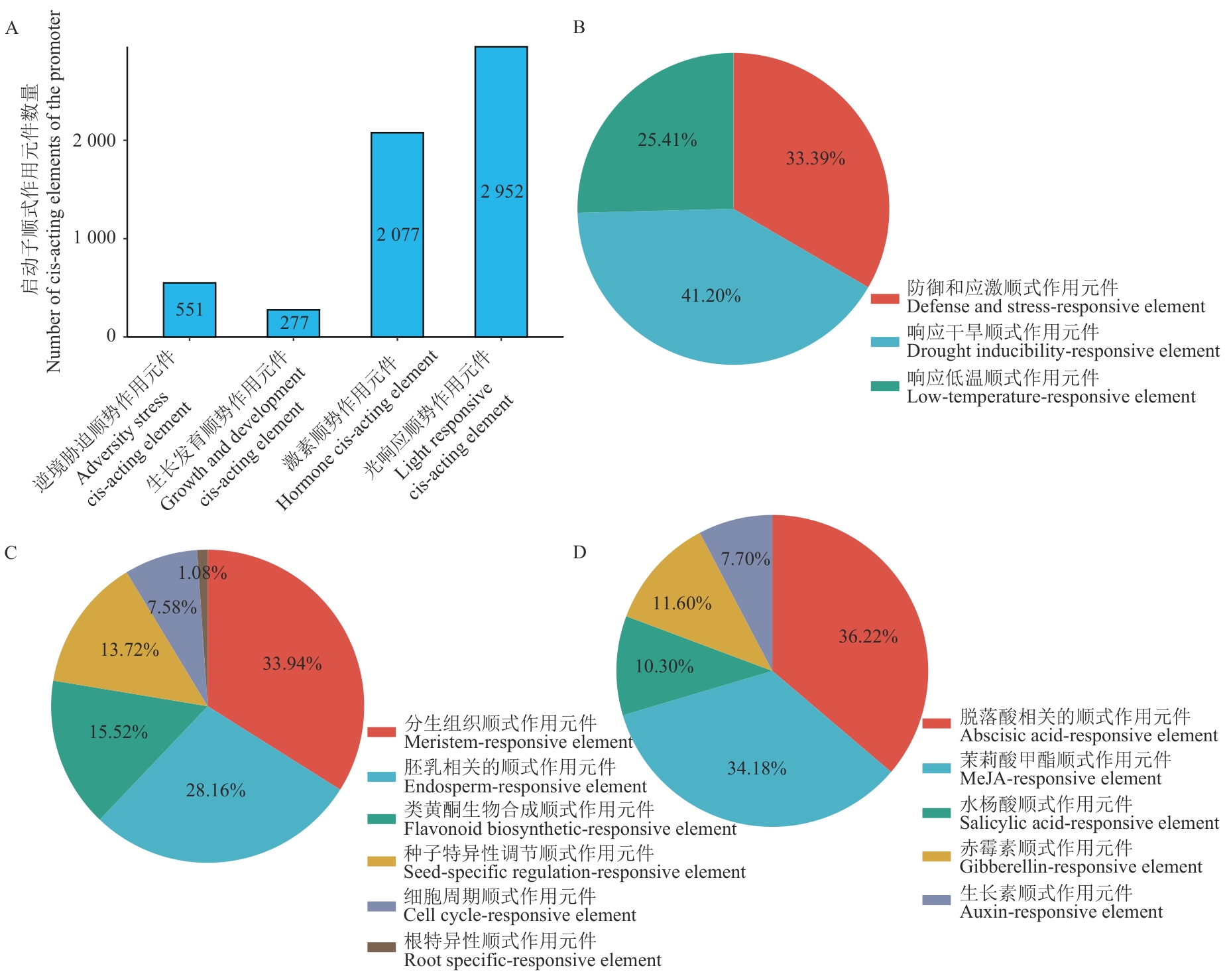
Fig. 5 Analysis of cis-acting elements in the promoters of soybean R2R3-MYB gene familyA: Number of cis-acting elements in different categories of promoters. B: cis-acting elements related to stress. C: cis-acting elements related to growth and development. D: cis-acting elements related to hormones
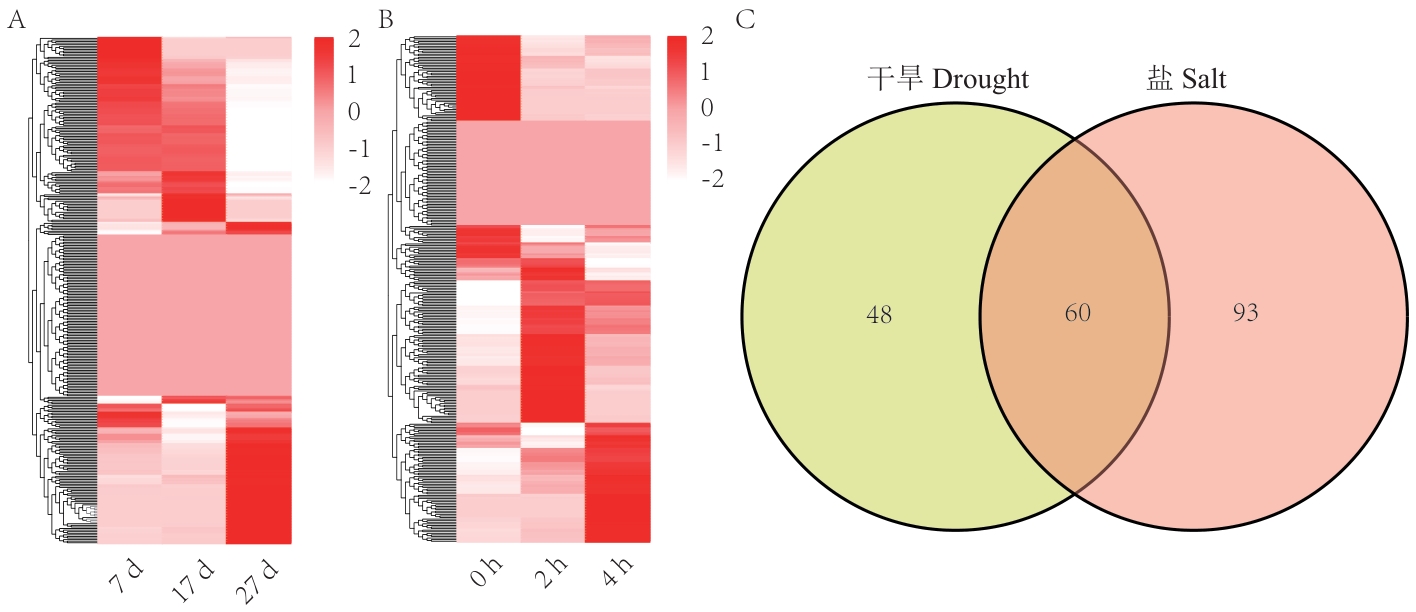
Fig. 6 Expression analysis of genes in soybean R2R3-MYB gene family under drought and salt stressA: Expression calorimetry under drought stress. B: Expression calorimetry under salt stress. C: Number of common and specific differentially expressed R2R3-MYB genes under drought and salt stress
| 17 | Zhang C, Xie L, Yu H, et al. The T2T genome assembly of soybean cultivar ZH13 and its epigenetic landscapes [J]. Mol Plant, 2023, 16(11): 1715-1718. |
| 18 | Lu SN, Wang JY, Chitsaz F, et al. CDD/SPARCLE: the conserved domain database in 2020 [J]. Nucleic Acids Res, 2020, 48(D1): D265-D268. |
| 19 | Mariethoz J, Alocci D, Gastaldello A, et al. Glycomics@ExPASy: bridging the gap [J]. Mol Cell Proteomics, 2018, 17(11): 2164-2176. |
| 20 | Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11 [J]. Mol Biol Evol, 2021, 38(7): 3022-3027. |
| 21 | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| 22 | Chen SF, Zhou YQ, Chen YR, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor [J]. Bioinformatics, 2018, 34(17): i884-i890. |
| 23 | Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown [J]. Nat Protoc, 2016, 11(9): 1650-1667. |
| 24 | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method [J]. Methods, 2001, 25(4): 402-408. |
| 25 | 张丹, 郑平, 邓彪, 等. 百香果(Passiflora edulis)R2R3-MYB基因家族的结构和功能分析 [J]. 果树学报, 2024, 41(1): 12-29. |
| Zhang D, Zheng P, Deng B, et al. Structure and function analysis of R2R3-MYB gene family from passion fruit(Passiflora edulis) [J]. J Fruit Sci, 2024, 41(1): 12-29. | |
| 26 | 郭玉婷, 杜长霞. 黄瓜R2R3-MYB亚家族鉴定及生物信息学分析[J]. 浙江农林大学学报, 2024, 41 (2): 286-296. |
| Guo YT, Du CX. Identification and bioinformatics analysis of R2R3-MYB subfamily in cucumber [J]. J Zhejiang A F Univ, 2024, 41 (2): 286-296. | |
| 1 | Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis [J]. Trends Plant Sci, 2010, 15(10): 573-581. |
| 2 | 薛英喜, 魏建华, 姜廷波, 等. 植物次生生长相关MYB转录因子研究进展 [J]. 安徽农业科学, 2012, 40(13): 7650-7655. |
| Xue YX, Wei JH, Jiang TB, et al. Research advances in the secondary growth-associated MYB transcription factors in plants [J]. J Anhui Agric Sci, 2012, 40(13): 7650-7655. | |
| 3 | 胡月, 王志富, 曹心如. 植物MYB转录因子的功能研究进展 [J]. 安徽农学通报, 2023, 29(12): 60-62. |
| Hu Y, Wang ZF, Cao XR. Progress in the functional study of plant MYB transcription factors [J]. Anhui Agric Sci Bull, 2023, 29(12): 60-62. | |
| 4 | Wilkins O, Nahal H, Foong J, et al. Expansion and diversification of the Populus R2R3-MYB family of transcription factors [J]. Plant Physiol, 2009, 149(2): 981-993. |
| 5 | Abdullah-Zawawi MR, Ahmad-Nizammuddin NF, Govender N, et al. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice [J]. Sci Rep, 2021, 11(1): 19678. |
| 6 | Wei QH, Chen R, Wei X, et al. Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344 [J]. BMC Genomics, 2020, 21(1): 792. |
| 7 | 胡雅丹, 伍国强, 刘晨, 等. MYB转录因子在调控植物响应逆境胁迫中的作用[J]. 生物技术通报, 2024, 40 (6): 5-22. |
| Hu YD, Wu GQ, Liu C, et al. Roles of MYB transcription factors in regulating the of plants to stress [J]. Biol Bull, 2024, 40 (6): 5-22. | |
| 8 | 令狐斌. 苦荞黄酮醇代谢转录因子FtMYB7和FtMYB9基因克隆及功能分析 [D]. 太谷: 山西农业大学, 2016. |
| Linghu B. Gene cloning and functional analysis of flavonoids alcohol metabolism transcription factors FtMYB7 and FtMYB9 in Fagopyrum tataricum Gaertn [D]. Taigu: Shanxi Agricultural University, 2016. | |
| 27 | 张超凡. MYB基因家族的鉴定及其在开花植物中的进化分析 [D]. 武汉: 华中农业大学, 2022. |
| Zhang CF. Identification of MYB gene family and its evolutionary analysis in flowering plants [D]. Wuhan: Huazhong Agricultural University, 2022. | |
| 28 | 梁细妹, 秦双双, 韦范, 等. 转录因子在植物干旱应激中的功能研究进展 [J]. 生物资源, 2024, 46(3): 220-230. |
| Liang XM, Qin SS, Wei F, et al. Research progress on the function of transcription factors in plant drought stress [J]. Biotic Resour, 2024, 46(3): 220-230. | |
| 29 | 潘志立, 郭雯, 王婷, 等. 叶片吸收水分的研究进展 [J]. 植物生理学报, 2021, 57(1): 19-32. |
| Pan ZL, Guo W, Wang T, et al. Research progress on foliar water uptake [J]. Plant Physiol J, 2021, 57(1): 19-32. | |
| 30 | 刘晓龙, 徐晨, 邵勤, 等. 脱落酸提高水稻抗逆性的研究进展 [J]. 东北农业科学, 2022, 47(6): 29-33. |
| Liu XL, Xu C, Shao Q, et al. Research progress of abscisic acid in improving rice stress resistance [J]. J Northeast Agric Sci, 2022, 47(6): 29-33. | |
| 31 | Daszkowska-Golec A, Szarejko I. Open or close the gate - stomata action under the control of phytohormones in drought stress conditions [J]. Front Plant Sci, 2013, 4: 138. |
| 32 | Mohamed HI, Latif HH. Improvement of drought tolerance of soybean plants by using methyl jasmonate [J]. Physiol Mol Biol Plants, 2017, 23(3): 545-556. |
| 33 | Sheteiwy MS, Shao HB, Qi WC, et al. Seed priming and foliar application with jasmonic acid enhance salinity stress tolerance of soybean (Glycine max L.) seedlings [J]. J Sci Food Agric, 2021, 101(5): 2027-2041. |
| 34 | Wang FB, Ren XQ, Zhang F, et al. A R2R3-type MYB transcription factor gene from soybean, GmMYB12, is involved in flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis [J]. Plant Biotechnol Rep, 2019, 13(3): 219-233. |
| 9 | 李红强, 罗倩, 杨帅, 等. 过表达NtMYB4a基因增强烟草抗旱能力 [J]. 亚热带植物科学, 2022, 51(1): 13-18. |
| Li HQ, Luo Q, Yang S, et al. Over-expression of NtMYB4a gene enhances drought resistance of tobacco [J]. Subtrop Plant Sci, 2022, 51(1): 13-18. | |
| 10 | 伍翀. 黄芩MYB转录因子功能初步研究 [D]. 武汉: 武汉工业学院, 2012. |
| Wu C. Preliminary study on the function of MYB transcription factor in Scutellaria baicalensis Georgi [D]. Wuhan: Wuhan Institute of Technology, 2012. | |
| 11 | Fang Q, Wang XQ, Wang HY, et al. The poplar R2R3 MYB transcription factor PtrMYB94 coordinates with abscisic acid signaling to improve drought tolerance in plants [J]. Tree Physiol, 2020, 40(1): 46-59. |
| 12 | Zhang ZJ, Zhang L, Liu Y, et al. Identification and expression analysis of R2R3-MYB family genes associated with salt tolerance in Cyclocarya paliurus [J]. Int J Mol Sci, 2022, 23(7): 3429. |
| 13 | Abe H, Urao T, Ito T, et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling [J]. Plant Cell, 2003, 15(1): 63-78. |
| 14 | Zhang LC, Liu GX, Zhao GY, et al. Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis [J]. Plant Cell Physiol, 2014, 55(10): 1802-1812. |
| 15 | Zhang LC, Zhao GY, Xia C, et al. A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis [J]. J Exp Bot, 2012, 63(16): 5873-5885. |
| 16 | 刘赵, 沈海军, 周凌晨, 等. 可溶性大豆多糖的提取及在食品中的应用研究进展 [J]. 中国调味品, 2023, 48(6): 199-208. |
| Liu Z, Shen HJ, Zhou LC, et al. Research progress on the extraction of soluble soybean polysaccharides and their application in food [J]. China Condiment, 2023, 48(6): 199-208. | |
| 35 | Wang N, Zhang WX, Qin MY, et al. Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor [J]. Plant Cell Physiol, 2017, 58(10): 1764-1776. |
| 36 | Shi K, Liu J, Liang H, et al. An alfalfa MYB-like transcriptional factor MsMYBH positively regulates alfalfa seedling drought resistance and undergoes MsWAV3-mediated degradation [J]. J Integr Plant Biol, 2024, 66(4): 683-699. |
| 37 | Xiao LY, Shi YY, Wang R, et al. The transcription factor OsMYBc and an E3 ligase regulate expression of a K+ transporter during salt stress [J]. Plant Physiol, 2022, 190(1): 843-859. |
| [1] | ZHAO Jing, GUO Qian, LI Rui-qi, LEI Ying-yang, YUE Ai-qin, ZHAO Jin-zhong, YIN Cong-cong, DU Wei-jun, NIU Jing-ping. Sequence Analysis and Induced Expression Analysis of GmGST Gene Cluster Genes in Soybean [J]. Biotechnology Bulletin, 2025, 41(5): 129-140. |
| [2] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [3] | SUN Tian-guo, YI Lan, QIN Xu-yang, QIAO Meng-xue, GU Xin-ying, HAN Yi, SHA Wei, ZHANG Mei-juan, MA Tian-yi. Genome-wide Identification of the DABB Gene Family in Brassica rapa ssp. pekinensis and Expression Analysis under Saline and Alkali Stress [J]. Biotechnology Bulletin, 2025, 41(4): 156-165. |
| [4] | WANG Tian-tian, CHANG Xue-rui, HUANG Wan-yang, HUANG Jia-xin, MIAO Ru-yi, LIANG Yan-ping, WANG Jing. Identification and Analysis of GASA Gene Family in Pepper (Capsicum annuum L.) [J]. Biotechnology Bulletin, 2025, 41(4): 166-175. |
| [5] | HUANG Jin-heng, HUANG Xi, ZHANG Jia-yan, ZHOU Xin-yu, LIAO Pei-ran, YANG Quan. Identification and Expression Analysis of the C3H Gene Family in Grona styracifolia across Different Varieties [J]. Biotechnology Bulletin, 2025, 41(4): 243-255. |
| [6] | BAN Qiu-yan, ZHAO Xin-yue, CHI Wen-jing, LI Jun-sheng, WANG Qiong, XIA Yao, LIANG Li-yun, HE Wei, LI Ye-yun, ZHAO Guang-shan. Cloning of Phytochrome Interaction Factor CsPIF3a and Its Response to Light and Temperature Stress in Camellia sinensis [J]. Biotechnology Bulletin, 2025, 41(4): 256-265. |
| [7] | LU Yong-jie, XIA Hai-qian, LI Yong-ling, ZHANG Wen-jian, YU Jing, ZHAO Hui-na, WANG Bing, XU Ben-bo, LEI Bo. Cloning and Expression Analysis of AP2/ERF Transcription Factor NtESR2 in Nicotiana tabacum [J]. Biotechnology Bulletin, 2025, 41(4): 266-277. |
| [8] | QIN Yue, YANG Yan, ZHANG Lei, LU Li-li, LI Xian-ping, JIANG Wei. Identification and Comparative Analysis of the StGAox Genes in Diploid and Tetraploid Potatoes [J]. Biotechnology Bulletin, 2025, 41(3): 146-160. |
| [9] | WANG Chen, LIU Guo-mei, CHEN Chang, ZHANG Jin-long, YAO Lin, SUN Xuan, DU Chun-fang. Genome-wide Identification and Expression Analysis of CCDs Family in Brassia rapa L. [J]. Biotechnology Bulletin, 2025, 41(3): 161-170. |
| [10] | MA Tian-yi, XU Jia-jia, LU Wen-jing, WU Yan, SHA Wei, ZHANG Mei-juan, PENG Yi-fang. Expression Analysis and Resistance Identification of BrcGASA3 in Chinese Cabbage ‘Jinxiaotong’ Cultivar under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 127-138. |
| [11] | XU Yuan-meng, MAO Jiao, WANG Meng-yao, WANG Shu, REN Jiang-ling, LIU Yu-han, LIU Si-chen, QIAO Zhi-jun, WANG Rui-yun, CAO Xiao-ning. Cloning and Expression Characteristics Analysis of Millet Genes PmDEP1 and PmEP3 [J]. Biotechnology Bulletin, 2025, 41(2): 150-162. |
| [12] | JIA Zi-jian, WANG Bao-qiang, CHEN Li-fei, WANG Yi-zhen, WEI Xiao-hong, ZHAO Ying. Expression Patterns of CHX Gene Family in Quinoa in Response to NO under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 163-174. |
| [13] | QIAN Zheng-yi, WU Shao-fang, CAO Shu-yi, SONG Ya-xin, PAN Xin-feng, LI Zhao-wei, FAN Kai. Identification of the NAC Transcription Factors in Nymphaea colorata and Their Expression Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 234-247. |
| [14] | HUANG Ying, YU Wen-jing, LIU Xue-feng, DIAO Gui-ping. Bioinformatics and Expression Pattern Analysis of Glutathione S-transferase in Populus davidiana × P. bolleana [J]. Biotechnology Bulletin, 2025, 41(2): 248-256. |
| [15] | XIANG Chun-fan, LI Le-song, WANG Juan, LIANG Yan-li, YANG Sheng-chao, LI Meng-fei, ZHAO Yan. Functional Identification and Expression Analysis of Cinnamonyl Alcohol Dehydrogenase AsCAD in Angelica sinensis [J]. Biotechnology Bulletin, 2025, 41(2): 295-308. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||