








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (6): 85-96.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1174
Previous Articles Next Articles
TANG Hao( ), SUN Can, LI Yuan-qiu, LUO Chao-bing(
), SUN Can, LI Yuan-qiu, LUO Chao-bing( )
)
Received:2020-09-18
Online:2021-06-26
Published:2021-07-08
Contact:
LUO Chao-bing
E-mail:tanghao89@aliyun.com;13366181512@163.com
TANG Hao, SUN Can, LI Yuan-qiu, LUO Chao-bing. Screening and Genome Sequencing of Cellulytic Bacterium Raoultella ornithinolytica LL1[J]. Biotechnology Bulletin, 2021, 37(6): 85-96.
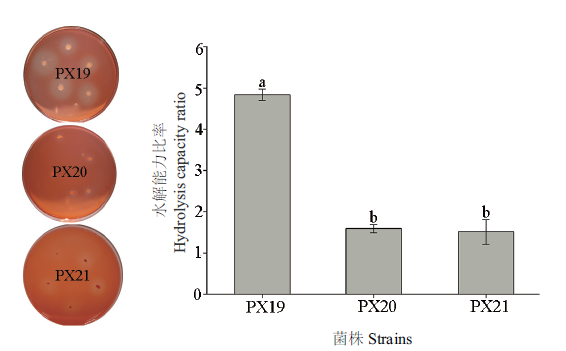
Fig.1 Comparison of Congo red staining of cellulose degrading bacterium PX19, PX20 and PX21 The average±standard error is shown in the figure, and different letters show significant difference in hydrolysis ability of different strains ( P < 0.05, Duncan’s test ). The same below
| 特征 Characteristics | 染色体 Chromosome | 质粒 Plasmid |
|---|---|---|
| 基因组大小Genome size/bp | 5 440 254 | 144 100 |
| GC含量GC content/% | 55.23 | 52.06 |
| 基因区域GC含量 GC content in gene region/% | 56.39 | 56.16 |
| 基因间区GC含量 GC content in gene spacer region/% | 48.63 | 55.13 |
| 基因数量 Gene number | 5 512 | 5 512 |
| 基因总长度Total gene length/bp | 4 687 047 | 102 264 |
| 重复序列 Repetitive sequence | 149 | 4 |
| 5S rRNA | 9 | - |
| 16S rRNA | 8 | - |
| 23S rRNA | 8 | - |
| tRNA | 87 | - |
| NR注释 NR annotation | 5 511 | - |
| Swiss-Prot注释 Swiss-Prot annotation | 4 419 | - |
| Pfam注释 Pfam annotation | 4 584 | - |
| COG注释 COG annotation | 4 745 | - |
| GO注释 Go annotation | 3 610 | - |
| KEGG注释 KEGG annotation | 3 336 | - |
Table1 Genome characteristics of R. ornithinolytica LL1
| 特征 Characteristics | 染色体 Chromosome | 质粒 Plasmid |
|---|---|---|
| 基因组大小Genome size/bp | 5 440 254 | 144 100 |
| GC含量GC content/% | 55.23 | 52.06 |
| 基因区域GC含量 GC content in gene region/% | 56.39 | 56.16 |
| 基因间区GC含量 GC content in gene spacer region/% | 48.63 | 55.13 |
| 基因数量 Gene number | 5 512 | 5 512 |
| 基因总长度Total gene length/bp | 4 687 047 | 102 264 |
| 重复序列 Repetitive sequence | 149 | 4 |
| 5S rRNA | 9 | - |
| 16S rRNA | 8 | - |
| 23S rRNA | 8 | - |
| tRNA | 87 | - |
| NR注释 NR annotation | 5 511 | - |
| Swiss-Prot注释 Swiss-Prot annotation | 4 419 | - |
| Pfam注释 Pfam annotation | 4 584 | - |
| COG注释 COG annotation | 4 745 | - |
| GO注释 Go annotation | 3 610 | - |
| KEGG注释 KEGG annotation | 3 336 | - |
| 序号 Serial number | COG类型 COG type | 基因数目 Number of genes | 相对比例 Relative ratio/% |
|---|---|---|---|
| 1 | 未知功能 Unknown function | 1 217 | 25.65 |
| 2 | 氨基酸转运与代谢 Amino acid transport and metabolism | 464 | 9.78 |
| 3 | 碳水化合物转运和代谢 Carbohydrate transport and metabolism | 454 | 9.57 |
| 4 | 转录 Transcription | 419 | 8.83 |
| 5 | 无机离子转运与代谢 Transport and metabolism of inorganic ions | 359 | 7.57 |
| 6 | 复制、重组和修复 Replication, recombination and repair | 313 | 6.60 |
| 7 | 能源生产与转换 Energy production and conversion | 274 | 5.77 |
| 8 | 细胞壁/膜/包膜生物发生 Cell wall / membrane / capsule biogenesis | 249 | 5.25 |
| 9 | 翻译、核糖体结构与生物发生 Translation, ribosomal structure and biogenesis | 189 | 3.98 |
| 10 | 翻译后修饰、蛋白质转运、分子伴侣 Post-translational modifications, protein transport, molecular chaperones | 151 | 3.18 |
| 11 | 信号转导机制 Signaling mechanism | 146 | 3.08 |
| 12 | 辅酶转运与代谢 Coenzyme transport and metabolism | 141 | 2.97 |
| 13 | 脂质转运与代谢 Lipid transport and metabolism | 94 | 1.98 |
| 14 | 核苷酸转运和代谢 Nucleotide transport and metabolism | 90 | 1.90 |
| 15 | 细胞内转运、分泌和囊泡转运 Intracellular transport, secretion and vesicle transport | 86 | 1.81 |
| 16 | 次生代谢物生物合成、运输和分解代谢 Biosynthesis, transport and catabolism of secondary metabolites | 73 | 1.54 |
| 17 | 防御机制 Defense mechanism | 55 | 1.16 |
| 18 | 细胞周期控制、细胞分裂和染色体分离 Cell cycle control, cell division and chromosome separation | 37 | 0.78 |
| 19 | 细胞运动性 Cell mobility | 18 | 0.38 |
| 20 | RNA加工与修饰 RNA processing and modification | 1 | 0.02 |
| 21 | 染色质结构与动力学 Chromatin structure and kinetics | 1 | 0.02 |
Table 2 COG annotation of R. ornithinolytica LL1
| 序号 Serial number | COG类型 COG type | 基因数目 Number of genes | 相对比例 Relative ratio/% |
|---|---|---|---|
| 1 | 未知功能 Unknown function | 1 217 | 25.65 |
| 2 | 氨基酸转运与代谢 Amino acid transport and metabolism | 464 | 9.78 |
| 3 | 碳水化合物转运和代谢 Carbohydrate transport and metabolism | 454 | 9.57 |
| 4 | 转录 Transcription | 419 | 8.83 |
| 5 | 无机离子转运与代谢 Transport and metabolism of inorganic ions | 359 | 7.57 |
| 6 | 复制、重组和修复 Replication, recombination and repair | 313 | 6.60 |
| 7 | 能源生产与转换 Energy production and conversion | 274 | 5.77 |
| 8 | 细胞壁/膜/包膜生物发生 Cell wall / membrane / capsule biogenesis | 249 | 5.25 |
| 9 | 翻译、核糖体结构与生物发生 Translation, ribosomal structure and biogenesis | 189 | 3.98 |
| 10 | 翻译后修饰、蛋白质转运、分子伴侣 Post-translational modifications, protein transport, molecular chaperones | 151 | 3.18 |
| 11 | 信号转导机制 Signaling mechanism | 146 | 3.08 |
| 12 | 辅酶转运与代谢 Coenzyme transport and metabolism | 141 | 2.97 |
| 13 | 脂质转运与代谢 Lipid transport and metabolism | 94 | 1.98 |
| 14 | 核苷酸转运和代谢 Nucleotide transport and metabolism | 90 | 1.90 |
| 15 | 细胞内转运、分泌和囊泡转运 Intracellular transport, secretion and vesicle transport | 86 | 1.81 |
| 16 | 次生代谢物生物合成、运输和分解代谢 Biosynthesis, transport and catabolism of secondary metabolites | 73 | 1.54 |
| 17 | 防御机制 Defense mechanism | 55 | 1.16 |
| 18 | 细胞周期控制、细胞分裂和染色体分离 Cell cycle control, cell division and chromosome separation | 37 | 0.78 |
| 19 | 细胞运动性 Cell mobility | 18 | 0.38 |
| 20 | RNA加工与修饰 RNA processing and modification | 1 | 0.02 |
| 21 | 染色质结构与动力学 Chromatin structure and kinetics | 1 | 0.02 |
| 序号 Serial number | 通路ID Pathway ID | 通路 Pathways | 基因数目 Number of genes | 相对比例 Relative proportion/% |
|---|---|---|---|---|
| 1 | ko00500 | 淀粉和蔗糖代谢 Starch and sucrose metabolism | 67 | 2.01 |
| 2 | ko00010 | 糖酵解/糖异生 Glycolysis/gluconeogenesis | 62 | 1.86 |
| 3 | ko00620 | 丙酮酸代谢 Pyruvate metabolism | 58 | 1.74 |
| 4 | ko00030 | 戊糖磷酸途径 Pentose phosphate pathway | 48 | 1.44 |
| 5 | ko00520 | 氨基糖和核苷酸糖代谢 Amino and nucleotide sugar metabolism | 47 | 1.41 |
| 6 | ko00051 | 果糖和甘露糖代谢 Fructose and mannose metabolism | 43 | 1.29 |
| 7 | ko00650 | 丁酸代谢 Butyric acid metabolism | 40 | 1.20 |
| 8 | ko00040 | 戊糖、葡萄糖醛酸转换 Conversion of pentose to glucuronic acid | 36 | 1.08 |
| 9 | ko00640 | 丙酸代谢 Propionic acid metabolism | 33 | 0.99 |
| 10 | ko00630 | 乙醛酸和二羧酸代谢 Glyoxylic acid and dicarboxylic acid metabolism | 33 | 0.99 |
| 11 | ko00020 | 三羧酸循环 Tricarboxylic acid cycle | 33 | 0.99 |
| 12 | ko00053 | 抗坏血酸和醛酸代谢 Ascorbic acid and aldehyde acid metabolism | 32 | 0.96 |
| 13 | ko00052 | 半乳糖代谢 Galactose metabolism | 26 | 0.78 |
| 14 | ko00660 | C5支链二元酸代谢 C5 branched diacid metabolism | 13 | 0.39 |
| 15 | ko00562 | 磷酸肌醇代谢Phosphoinositide metabolism | 11 | 0.33 |
Table 3 KEGG pathways related to carbohydrate metabolism
| 序号 Serial number | 通路ID Pathway ID | 通路 Pathways | 基因数目 Number of genes | 相对比例 Relative proportion/% |
|---|---|---|---|---|
| 1 | ko00500 | 淀粉和蔗糖代谢 Starch and sucrose metabolism | 67 | 2.01 |
| 2 | ko00010 | 糖酵解/糖异生 Glycolysis/gluconeogenesis | 62 | 1.86 |
| 3 | ko00620 | 丙酮酸代谢 Pyruvate metabolism | 58 | 1.74 |
| 4 | ko00030 | 戊糖磷酸途径 Pentose phosphate pathway | 48 | 1.44 |
| 5 | ko00520 | 氨基糖和核苷酸糖代谢 Amino and nucleotide sugar metabolism | 47 | 1.41 |
| 6 | ko00051 | 果糖和甘露糖代谢 Fructose and mannose metabolism | 43 | 1.29 |
| 7 | ko00650 | 丁酸代谢 Butyric acid metabolism | 40 | 1.20 |
| 8 | ko00040 | 戊糖、葡萄糖醛酸转换 Conversion of pentose to glucuronic acid | 36 | 1.08 |
| 9 | ko00640 | 丙酸代谢 Propionic acid metabolism | 33 | 0.99 |
| 10 | ko00630 | 乙醛酸和二羧酸代谢 Glyoxylic acid and dicarboxylic acid metabolism | 33 | 0.99 |
| 11 | ko00020 | 三羧酸循环 Tricarboxylic acid cycle | 33 | 0.99 |
| 12 | ko00053 | 抗坏血酸和醛酸代谢 Ascorbic acid and aldehyde acid metabolism | 32 | 0.96 |
| 13 | ko00052 | 半乳糖代谢 Galactose metabolism | 26 | 0.78 |
| 14 | ko00660 | C5支链二元酸代谢 C5 branched diacid metabolism | 13 | 0.39 |
| 15 | ko00562 | 磷酸肌醇代谢Phosphoinositide metabolism | 11 | 0.33 |
| 分类 Classification | 基因ID Gene ID | 功能预测 Function prediction |
|---|---|---|
| 纤维素降解相关基因 Cellulose degradation related genes | gene2786 | GH9 (endoglucanase EC 3.2.1.4) |
| gene0210 | GH8 (endoglucanase EC 3.2.1.4) | |
| gene0218 | GH8 (endoglucanase EC 3.2.1.4) | |
| gene0074 | GH4 (6-phospho-beta-glucosidase EC 3.2.1.86) | |
| gene3342 | GH4 (6-phospho-beta-glucosidase EC 3.2.1.86) | |
| gene1935 | GH4 (alpha-glucosidase EC 3.2.1.21) | |
| gene1744 | GH3 (beta-glucosidase EC 3.2.1.21) | |
| gene0827 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene0964 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1208 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1476 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1736 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1922 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene2493 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene2777 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene3805 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1047 | GH32 (sucrose-6-phosphate hydrolase EC 2.4.1.-) | |
| gene0301 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene0164 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene4341 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene4934 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene3011 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene2026 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene0300 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene3799 | GH31 (α-glucosidase EC 3.2.1.20) | |
| 半纤维素降解相关基因 Hemicellulose degradation related genes | gene1732 | GH43 (xylan 1,4-beta-xylosidase EC:3.2.1.37) |
| gene4854 | GH43 (xylan 1,4-beta-xylosidase EC:3.2.1.37) | |
| gene4896 | GH43 (xylan 1,4-beta-xylosidase EC:3.2.1.37) | |
| gene4257 | GH53 (arabinogalactan endo-1,4-beta-galactosidase EC:3.2.1.89) | |
| gene4256 | GH42 (beta-galactosidase EC:3.2.1.23) | |
| gene2601 | GH2 (beta-galactosidase EC:3.2.1.23) | |
| 木质素降解相关基因 Lignin degradation related genes | gene1352 | AA1 (laccase EC 1.10.3.2) |
| gene2367 | AA1 (laccase EC 1.10.3.2) | |
| gene2810 | AA2 (manganese peroxidase EC 1.11.1.13) | |
| gene0833 | AA7 (FAD-binding protein) | |
| gene0243 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene1369 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene2549 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene3520 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene3990 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene4124 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene4303 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) |
Table 4 Genes encoding lignocellulose degradation in R. ornithinolytica LL1
| 分类 Classification | 基因ID Gene ID | 功能预测 Function prediction |
|---|---|---|
| 纤维素降解相关基因 Cellulose degradation related genes | gene2786 | GH9 (endoglucanase EC 3.2.1.4) |
| gene0210 | GH8 (endoglucanase EC 3.2.1.4) | |
| gene0218 | GH8 (endoglucanase EC 3.2.1.4) | |
| gene0074 | GH4 (6-phospho-beta-glucosidase EC 3.2.1.86) | |
| gene3342 | GH4 (6-phospho-beta-glucosidase EC 3.2.1.86) | |
| gene1935 | GH4 (alpha-glucosidase EC 3.2.1.21) | |
| gene1744 | GH3 (beta-glucosidase EC 3.2.1.21) | |
| gene0827 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene0964 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1208 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1476 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1736 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1922 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene2493 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene2777 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene3805 | GH1 (beta-glucosidase EC 3.2.1.21) | |
| gene1047 | GH32 (sucrose-6-phosphate hydrolase EC 2.4.1.-) | |
| gene0301 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene0164 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene4341 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene4934 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene3011 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene2026 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene0300 | GH13 (α-glucosidase EC 3.2.1.20) | |
| gene3799 | GH31 (α-glucosidase EC 3.2.1.20) | |
| 半纤维素降解相关基因 Hemicellulose degradation related genes | gene1732 | GH43 (xylan 1,4-beta-xylosidase EC:3.2.1.37) |
| gene4854 | GH43 (xylan 1,4-beta-xylosidase EC:3.2.1.37) | |
| gene4896 | GH43 (xylan 1,4-beta-xylosidase EC:3.2.1.37) | |
| gene4257 | GH53 (arabinogalactan endo-1,4-beta-galactosidase EC:3.2.1.89) | |
| gene4256 | GH42 (beta-galactosidase EC:3.2.1.23) | |
| gene2601 | GH2 (beta-galactosidase EC:3.2.1.23) | |
| 木质素降解相关基因 Lignin degradation related genes | gene1352 | AA1 (laccase EC 1.10.3.2) |
| gene2367 | AA1 (laccase EC 1.10.3.2) | |
| gene2810 | AA2 (manganese peroxidase EC 1.11.1.13) | |
| gene0833 | AA7 (FAD-binding protein) | |
| gene0243 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene1369 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene2549 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene3520 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene3990 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene4124 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) | |
| gene4303 | AA3 (cellobiose dehydrogenase EC 1.1.99.18) |
| [1] | Himmel ME. Biomass recalcitrance:engineering plants and enzymes for biofuels production[J]. Science, 2007(5827):982. |
| [2] |
Bomble YJ, Lin CY, Amore A, et al. Lignocellulose deconstruction in the biosphere[J]. Curr Opin Chem Biol, 2017, 41:61-70.
doi: 10.1016/j.cbpa.2017.10.013 URL |
| [3] |
Pollegioni L, Tonin F, Rosini E. Lignin-degrading enzymes[J]. FEBS Journal, 2015, 282(7):1190-1213.
doi: 10.1111/febs.2015.282.issue-7 URL |
| [4] | Horn SJ, Vaaje-Kolstad G, Westereng B, et al. Novel enzymes for the degradation of cellulose[J]. Biotech Biof, 2012, 5:45. |
| [5] |
Karlsson J, Momcilovic D, et al. Enzymatic degradation of carboxymethyl cellulose hydrolyzed by the endoglucanases Cel5A, Cel7B, and Cel45A from Humicola insolens and Cel7B, Cel12A and Cel45Acore from Trichoderma reesei[J]. Biopolymers, 2002, 63(1):32-40.
pmid: 11754346 |
| [6] |
Hasper AA, Visser J, et al. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates D-xylose reductase gene expression[J]. Mol Microb, 2000, 36(1):193-200.
doi: 10.1046/j.1365-2958.2000.01843.x URL |
| [7] |
Du J, Zhang X, Li X, et al. The cellulose binding region in Trichoderma reesei cellobiohydrolase I has a higher capacity in improving crystalline cellulose degradation than that of Penicillium oxalicum[J]. Bioresource Technology, 2018, 266:19-25.
doi: 10.1016/j.biortech.2018.06.050 URL |
| [8] | Alonso-Pernas P, Bartram S, et al. Corrigendum: In vivo isotopic labeling of symbiotic bacteria involved in cellulose degradation and nitrogen recycling within the gut of the forest cockchafer(Melolontha hippocastani)[J]. Front Microbiol, 2018, 12:488. |
| [9] |
Ghosh D, Jana BB, Lahiri S, et al. Assessing the cellulase enzyme heterogeneity of bacterial strains and their feedback to cattle manure degradation in a greenhouse model of in vivo pond ecosystem[J]. Environmental Monitoring and Assessment, 2018, 190(8):452.
doi: 10.1007/s10661-018-6821-1 URL |
| [10] |
Oke MA, Annuar MSM, Simarani K. Mixed lignocellulosic biomass degradation and utilization for bacterial cellulase production[J]. Waste and Biomass Valorization, 2017, 8(3):893-903.
doi: 10.1007/s12649-016-9595-0 URL |
| [11] |
Li HH, Yu H, Sun MT, et al. Simultaneous removal and measurement of sulfide on the basis of turn-on fluorimetry[J]. Int J Environ Sci Technol, 2018, 15(6):1193-1200.
doi: 10.1007/s13762-017-1483-z URL |
| [12] |
Yadav S, Dubey SK. Cellulose degradation potential of Paenibacillus lautus strain BHU3 and its whole genome sequence[J]. Bioresource Technology, 2018, 262:124-131.
doi: 10.1016/j.biortech.2018.04.067 URL |
| [13] |
Kane SD, French CE. Characterisation of novel biomass degradation enzymes from the genome of Cellulomonas fimi[J]. Enzyme and Microbial Technology, 2018, 113:9-17.
doi: 10.1016/j.enzmictec.2018.02.004 URL |
| [14] | Wang S, Zhao D, Bai X, et al. Identification and characterization of a large protein essential for degradation of the crystalline region of cellulose by Cytophaga hutchinsonii[J]. Applied and Environmental Microbiology, 2016, 83(1):e02270-16. |
| [15] |
Lewin GR, Carlos C, Chevrette MG, et al. Evolution and ecology of Actinobacteria and their bioenergy applications[J]. Annual Review of Microbiology, 2016, 70:235-254.
doi: 10.1146/annurev-micro-102215-095748 URL |
| [16] | 黄胜威. 暗黑鳃金龟幼虫肠道微生物分子多态性及纤维素降解菌多样性研究[D]. 武汉:华中农业大学, 2012. |
| Huang S. Study on microbiota diversity and cellulolytica bacterial community in the gut of Holotrichia parallela larvae(Coleoptera:Scarabaeidae)[D]. Wuhan:Huazhong Agricultural University, 2012. | |
| [17] | 胡霞, 傅慧静, 李俊楠, 等. 松墨天牛幼虫肠道纤维素降解细菌的分离与鉴定[J]. 福建农林大学学报:自然科学版, 2018, 47(3):322-328. |
| Hu X, Fu H, Li J, et al. Isolation and identification of cellulolytic bacteria associated with the gut of Monochamus alternatus larvae[J]. Journal of Fujian Agriculture and Forestry University:Natural Science Edition, 2018, 47(3):322-328. | |
| [18] | 傅慧静. 松墨天牛肠道细菌多样性和粘质沙雷氏菌木质素降解特性的研究[D]. 福州:福建农林大学, 2017. |
| Fu H. Studies on bacteria diversity in the gut of Monochamus alternatus Hope and lignin-degradation characteristics of Serratia marcescen[D]. Fuzhou:Fujian Agriculture and Forestry University, 2017. | |
| [19] |
Luo C, Li Y, Liao H, et al. De novo transcriptome assembly of the bamboo snout beetle Cyrtotrachelus buqueti reveals ability to degrade lignocellulose of bamboo feedstock[J]. Biotechnology for Biofuels, 2018, 11:292-312.
doi: 10.1186/s13068-018-1291-9 URL |
| [20] | Luo C, Li Y, Chen Y, et al. Degradation of bamboo lignocellulose by bamboo snout beetle Cyrtotrachelus buqueti in vivo and vitro:efficiency and mechanism[J]. Biotech Biof, 2019, 12:75-89. |
| [21] |
Luo C, Li Y, Chen Y, et al. Bamboo lignocellulose degradation by gut symbiotic microbiota of the bamboo snout beetle Cyrtotrachelus buqueti[J]. Biotechnology for Biofuels, 2019, 12:70-86.
doi: 10.1186/s13068-019-1411-1 URL |
| [22] |
Li Y, Lei L, Zheng L, et al. Genome sequencing of gut symbiotic Bacillus velezensis LC1 for bioethanol production from bamboo shoots[J]. Biotechnology for Biofuels, 2020, 13:34.
doi: 10.1186/s13068-020-1671-9 URL |
| [23] |
Teather RM, Wood PJ. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from bovine rumen[J]. Applied and Environmental Microbiology, 1959, 43:777-80.
doi: 10.1128/aem.43.4.777-780.1982 URL |
| [24] | Motham M, Pumas C, Peerapornpisal Y. Improvement of DNA extraction protocols for Nostochopsis spp[J]. Chiang Mai Journal of Science, 2014, 41(3):557-567. |
| [25] |
Westbrook CJ, Karl JA, et al. No assembly required:full-length MHC class I allele discovery by PacBio circular consensus sequencing[J]. Hum Immunol, 2015, 76:891-896.
doi: 10.1016/j.humimm.2015.03.022 URL |
| [26] |
Genomic and secretomic insight into lignocellulolytic system of an endophytic bacterium Pantoea ananatis Sd-1[J]. Biotechnology for Biofuels, 2016, 9(1):25.
doi: 10.1186/s13068-016-0439-8 URL |
| [27] | Eddy RS. A new generation of homology search tools based on probabilistic inference[J]. Gen Infor, 2009:205-211. |
| [28] |
Chen C, Chen H, Zhang Y, et al. TBtools-an integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [29] | Bao W, Zhou Y, Jiang J, et al. Complete genome sequence of Raoultella ornithinolytica strain S12, a lignin-degrading bacterium isolated from forest soil[J]. Gen Ann, 2015, 3(2):e00104-15. |
| [30] | Shin SH, Um Y, Beak JH, et al. Complete genome sequence of Raoultella ornithinolytica strain B6, a 2, 3-butanediol-producing bacterium isolated from oil-contaminated soil[J]. Genome Announcement, 2013, 1(3):e00395-13. |
| [31] |
Kim T, Cho S, Lee SM, et al. High production of 2, 3-butanediol(2, 3-BD)by Raoultella ornithinolytica B6 via optimizing fermentation conditions and overexpressing 2, 3-BD Synjournal Genes[J]. PLoS One, 2016, 11(10):e0165076.
doi: 10.1371/journal.pone.0165076 URL |
| [32] |
Kim T, Cho S, Woo HM, et al. High production of 2, 3-butanediol from glycerol without 1, 3-propanediol formation by Raoultella ornithinolytica B6[J]. Appl Microbiol Biotechnol, 2017, 101(7):2821-2830.
doi: 10.1007/s00253-017-8094-y URL |
| [33] | Roth C, Weizenmann N, Bexten N, et al. Amylose recognition and ring-size determination of amylomaltase[J]. Science Advances, 2017, 3:1360-1386. |
| [34] |
van den Brink J, de Vries RP. Fungal enzyme sets for plant polysaccharide degradation[J]. Applied Microbiology and Biotechnology, 2011, 91(6):1477-1492.
doi: 10.1007/s00253-011-3473-2 URL |
| [35] |
Machida M, Asai K, et al. Genome sequencing and analysis of Aspergillus oryzae[J]. Nature, 2005, 438(7071):1157-1161.
doi: 10.1038/nature04300 URL |
| [36] |
Vanholme B, Jacob J, Cannoot B, et al. Arabinogalacta endo-1, 4-β-galactosidase:a putative plant cell wall-degrading enzyme of plant-parasitic nematodes[J]. Nematology, 2009, 11:739-747.
doi: 10.1163/156854109X404599 URL |
| [37] | Zhang J, Siika-Aho M, Tenkanen M, et al. The role of acetyl xylan esterase in the solubilization of xylan and enzymatic hydrolysis of wheat straw and giant reed[J]. Biotech Biof, 2011, 4:60-81. |
| [38] |
Mamo G, Hatti-Kaul R, Bo M. A thermostable alkaline active endo-β-1-4-xylanase from Bacillus halodurans S7:purification and characterization[J]. Enzyme and Microbial Technology, 2006, 39:1492-1498.
doi: 10.1016/j.enzmictec.2006.03.040 URL |
| [39] |
Biely P. Microbial carbohydrate esterases deacetylating plant polysaccharides[J]. Biotech Adv, 2012, 30:1575-1588.
doi: 10.1016/j.biotechadv.2012.04.010 URL |
| [40] |
Zhao Z, Liu H, Wang C, et al. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi[J]. BMC Genom, 2014, 15:6-12.
doi: 10.1186/1471-2164-15-6 URL |
| [41] | Van den Heuvel RH, Fraaije MW, Mattevi A, et al. Structure, function and redesign of vanillyl-alcohol oxidase[J]. International Congress, 2002, 1233:13-24. |
| [42] |
Levasseur A, Drula E, Lombard V, et al. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes[J]. Biotechnology for Biofuels, 2013, 6:41-52.
doi: 10.1186/1754-6834-6-41 URL |
| [1] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [2] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [3] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [4] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [5] | ZHANG Jing, ZHANG Hao-rui, CAO Yun, HUANG Hong-ying, QU Ping, ZHANG Zhi-ping. Research Progress in Thermophilic Microorganisms for Cellulose Degradation [J]. Biotechnology Bulletin, 2023, 39(6): 73-87. |
| [6] | MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose [J]. Biotechnology Bulletin, 2023, 39(4): 124-135. |
| [7] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| [8] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [9] | QU Chun-juan, ZHU Yue, JIANG Chen, QU Ming-jing, WANG Xiang-yu, LI Xiao. Whole Mitochondrial Genome and Phylogeny Analysis of Anomala corpulenta [J]. Biotechnology Bulletin, 2023, 39(2): 263-273. |
| [10] | ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101 [J]. Biotechnology Bulletin, 2023, 39(10): 292-303. |
| [11] | WANG Shuai, LV Hong-rui, ZHANG Hao, WU Zhan-wen, XIAO Cui-hong, SUN Dong-mei. Whole-Genome Sequencing Identification of Phosphate-solubilizing Bacteria PSB-R and Analysis of Its Phosphate-solubilizing Properties [J]. Biotechnology Bulletin, 2023, 39(1): 274-283. |
| [12] | WEN Chang, LIU Chen, LU Shi-yun, XU Zhong-bing, AI Chao-fan, LIAO Han-peng, ZHOU Shun-gui. Biological Characteristics and Genome Analysis of a Novel Multidrug-resistant Shigella flexneri Phage [J]. Biotechnology Bulletin, 2022, 38(9): 127-135. |
| [13] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [14] | ZHANG Kai-ping, LIU Yan-li, TU Mian-liang, LI Ji-wei, WU Wen-biao. Optimization of Producing Cellulase by Aspergillus fumigatus A-16 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(9): 215-225. |
| [15] | WANG Xin-guang, TIAN Lei, WANG En-ze, ZHONG Cheng, TIAN Chun-jie. Construction of Microbial Consortium for Efficient Degradation of Corn Straw and Evaluation of Its Degradation Effect [J]. Biotechnology Bulletin, 2022, 38(4): 217-229. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||