








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (1): 274-283.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0448
Previous Articles Next Articles
WANG Shuai1( ), LV Hong-rui1, ZHANG Hao1, WU Zhan-wen1, XIAO Cui-hong1, SUN Dong-mei1,2(
), LV Hong-rui1, ZHANG Hao1, WU Zhan-wen1, XIAO Cui-hong1, SUN Dong-mei1,2( )
)
Received:2022-04-12
Online:2023-01-26
Published:2023-02-02
Contact:
SUN Dong-mei
E-mail:15846761937@163.com;sdmlzw@126.com
WANG Shuai, LV Hong-rui, ZHANG Hao, WU Zhan-wen, XIAO Cui-hong, SUN Dong-mei. Whole-Genome Sequencing Identification of Phosphate-solubilizing Bacteria PSB-R and Analysis of Its Phosphate-solubilizing Properties[J]. Biotechnology Bulletin, 2023, 39(1): 274-283.
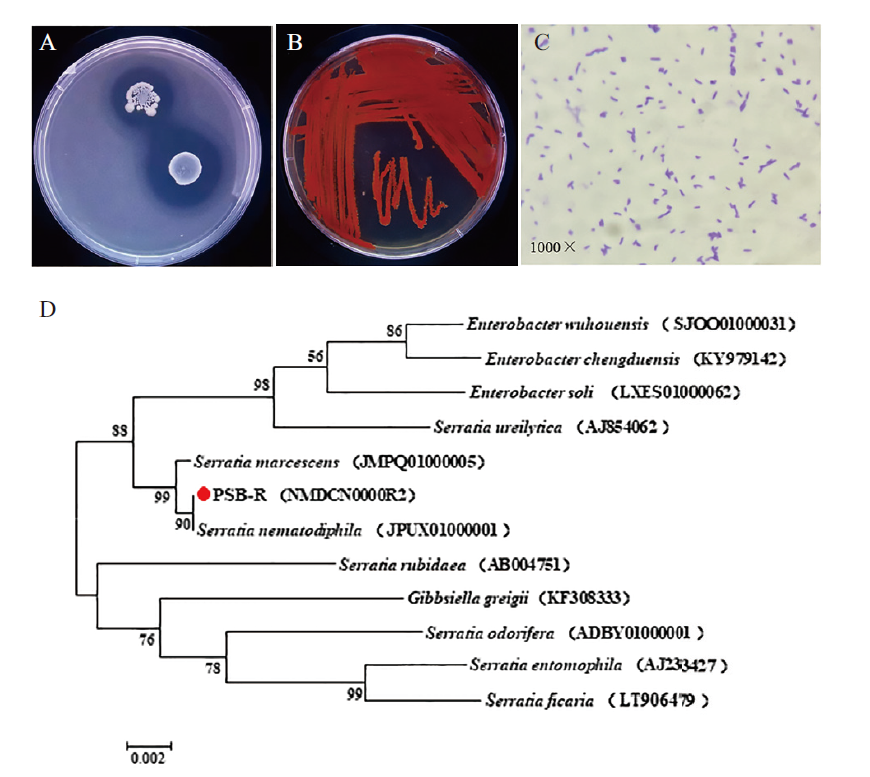
Fig. 1 Identification results of PSB-R A: Growth results of PSB-R on inorganic phosphorus medium. B: Growth results of PSB-R on nutrient agar surface. C: Morphology of PSB-R bacteria(crystal violet staining). D: Phylogenetic tree based on 16S rRNA gene sequence
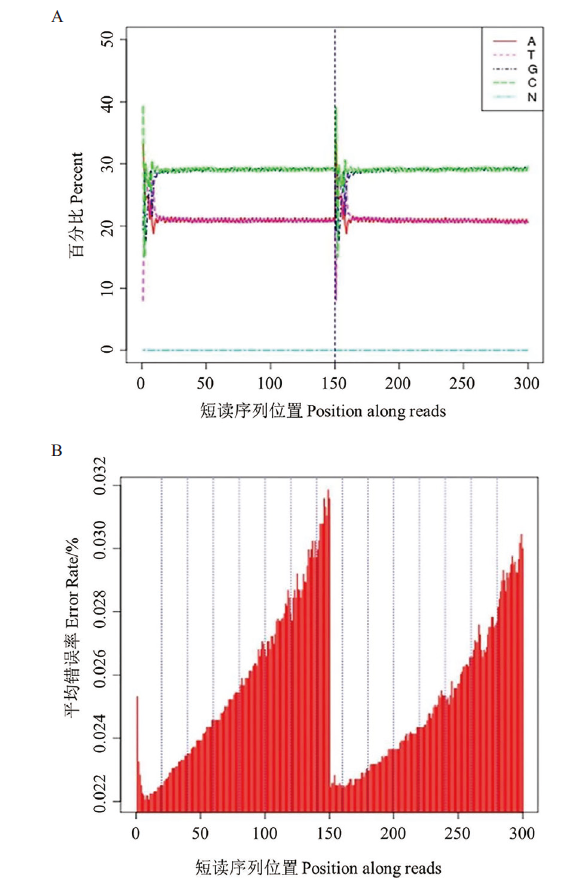
Fig. 2 Base content and mass distribution map A: Base content distribution map: the abscissa refers to the position of the reads where the base is located, and the ordinate refers to the content of each base at a certain position. B: Base mdistribution map: the abscissa refers to the position of the reads where the base is located, and the ordinate refers to the average error rate percentage at that position of the reads
| 项目Item | 数据Data |
|---|---|
| Scaffold数量 Number of Scaffold | 16 |
| Contig数量 Number of Contig | 16 |
| (G+C)/% | 59.75 |
| 基因组大小Genome size/bp | 5 061 510 |
| 预测基因数量 Number of genes | 4 742 |
| 基因总长度Total gene length/bp | 4 393 551 |
Table 1 Assembly statistics of genomes
| 项目Item | 数据Data |
|---|---|
| Scaffold数量 Number of Scaffold | 16 |
| Contig数量 Number of Contig | 16 |
| (G+C)/% | 59.75 |
| 基因组大小Genome size/bp | 5 061 510 |
| 预测基因数量 Number of genes | 4 742 |
| 基因总长度Total gene length/bp | 4 393 551 |
| 基因号码Gene code | 基因名称Gene name | 产物名称 Product name | 生物活性 Activity |
|---|---|---|---|
| PSB.R_GM000437 | gcd | PQQ依赖型葡萄糖脱氢酶 Quinoprotein glucose dehydrogenase | 葡萄糖酸合成 Gluconic acid synthesis |
| PSB.R GM001125 | ppq B | PQQ合成蛋白B Cofactor PQQ biosynthesis protein B | |
| PSB.R GM001126 | ppq C | PQQ合成蛋白C Cofactor PQQ biosynthesis protein C | |
| PSB.R GM001127 | ppq D | PQQ合成蛋白D Cofactor PQQ biosynthesis protein D | |
| PSB.R GM001128 | ppq E | PQQ合成蛋白E Cofactor PQQ biosynthesis protein E | |
| PSB.R GM001129 | ppq F | PQQ合成蛋白F Cofactor PQQ biosynthesis protein F | |
| PSB.R_GM000385 | ent A | 2,3-二氢-2,3-二羟基苯甲酸脱氢酶 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | 铁载体产生 Siderophore production |
| PSB.R_GM003128 | ent C | 异分支酸合成酶C Isochorismate synthase C | |
| PSB.R_GM003126 | ent B | 肠杆菌素合成亚基B Enterobactin synthase subunit B | |
| PSB.R_GM003127 | ent E | 肠杆菌素合成亚基E Enterobactin synthase subunit E | |
| PSB.R_GM003130 | ent F | 肠杆菌素合成亚基F Enterobactin synthase subunit F | |
| PSB.R_GM003129 | ent S | 肠杆菌素(铁载体)外排蛋白 Enterobactin(siderophore)exporter | |
| PSB.R_GM002907 | exb B | 生物聚合物转运蛋白B Biopolymer transport protein B | |
| PSB.R_GM002910 | exb D | 生物聚合物转运蛋白D Biopolymer transport protein D | |
| PSB.R_GM004736 | bfr | 细菌铁蛋白 Bacterioferritin | |
| PSB.R_GM000070 | ipd C | 吲哚丙酮酸脱羧酶 Indolepyruvate decarboxylase | IAA产生 IAA production |
| PSB.R_GM000012 | chi | 几丁质酶 Chitinase | 几丁质酶产生 Chitinase production |
| PSB.R_GM003955 | ppx | 外切聚磷酸酶 Exopolyphosphatase | 磷酸盐降解酶 Phosphate degrading enzyme |
| PSB.R_GM003047 | ppa | 无机焦磷酸酶 Inorganic pyrophosphatase | |
| PSB.R_GM001406 | phn X | 磷酸乙醛水解酶 Phosphonoacetaldehyde hydrolase | |
| PSB.R_GM002025 | pho A | 碱性磷酸酶 Alkaline phosphatase |
Table 2 Prediction of growth-promoting genes
| 基因号码Gene code | 基因名称Gene name | 产物名称 Product name | 生物活性 Activity |
|---|---|---|---|
| PSB.R_GM000437 | gcd | PQQ依赖型葡萄糖脱氢酶 Quinoprotein glucose dehydrogenase | 葡萄糖酸合成 Gluconic acid synthesis |
| PSB.R GM001125 | ppq B | PQQ合成蛋白B Cofactor PQQ biosynthesis protein B | |
| PSB.R GM001126 | ppq C | PQQ合成蛋白C Cofactor PQQ biosynthesis protein C | |
| PSB.R GM001127 | ppq D | PQQ合成蛋白D Cofactor PQQ biosynthesis protein D | |
| PSB.R GM001128 | ppq E | PQQ合成蛋白E Cofactor PQQ biosynthesis protein E | |
| PSB.R GM001129 | ppq F | PQQ合成蛋白F Cofactor PQQ biosynthesis protein F | |
| PSB.R_GM000385 | ent A | 2,3-二氢-2,3-二羟基苯甲酸脱氢酶 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | 铁载体产生 Siderophore production |
| PSB.R_GM003128 | ent C | 异分支酸合成酶C Isochorismate synthase C | |
| PSB.R_GM003126 | ent B | 肠杆菌素合成亚基B Enterobactin synthase subunit B | |
| PSB.R_GM003127 | ent E | 肠杆菌素合成亚基E Enterobactin synthase subunit E | |
| PSB.R_GM003130 | ent F | 肠杆菌素合成亚基F Enterobactin synthase subunit F | |
| PSB.R_GM003129 | ent S | 肠杆菌素(铁载体)外排蛋白 Enterobactin(siderophore)exporter | |
| PSB.R_GM002907 | exb B | 生物聚合物转运蛋白B Biopolymer transport protein B | |
| PSB.R_GM002910 | exb D | 生物聚合物转运蛋白D Biopolymer transport protein D | |
| PSB.R_GM004736 | bfr | 细菌铁蛋白 Bacterioferritin | |
| PSB.R_GM000070 | ipd C | 吲哚丙酮酸脱羧酶 Indolepyruvate decarboxylase | IAA产生 IAA production |
| PSB.R_GM000012 | chi | 几丁质酶 Chitinase | 几丁质酶产生 Chitinase production |
| PSB.R_GM003955 | ppx | 外切聚磷酸酶 Exopolyphosphatase | 磷酸盐降解酶 Phosphate degrading enzyme |
| PSB.R_GM003047 | ppa | 无机焦磷酸酶 Inorganic pyrophosphatase | |
| PSB.R_GM001406 | phn X | 磷酸乙醛水解酶 Phosphonoacetaldehyde hydrolase | |
| PSB.R_GM002025 | pho A | 碱性磷酸酶 Alkaline phosphatase |
| [1] | 王雪郦, 张芮瑞, 周少奇, 等. 不良环境解磷微生物研究进展[J]. 河南农业科学, 2020, 49(7): 8-17. |
| Wang XL, Zhang RR, Zhou SQ, et al. Research progress of phosphate-solubilizing microorganisms in bad environments[J]. J Henan Agric Sci, 2020, 49(7): 8-17. | |
| [2] | 吉蓉. 土壤解磷微生物及其解磷机制综述[J]. 甘肃农业科技, 2013(8): 42-45. |
| Ji R. Research summary on phosphate dissolution of phosphate solubilizing microorganisms[J]. Gansu Agric Sci Technol, 2013(8): 42-45. | |
| [3] |
Schindler DW, Hecky RE, Findlay DL, et al. Eutrophication of lakes cannot be controlled by reducing nitrogen input: results of a 37-year whole-ecosystem experiment[J]. Proc Natl Acad Sci USA, 2008, 105(32): 11254-11258.
doi: 10.1073/pnas.0805108105 pmid: 18667696 |
| [4] |
Boubekri K, Soumare A, Mardad I, et al. The screening of potassium- and phosphate-solubilizing actinobacteria and the assessment of their ability to promote wheat growth parameters[J]. Microorganisms, 2021, 9(3): 470.
doi: 10.3390/microorganisms9030470 URL |
| [5] | Whitelaw MA. Growth promotion of plants inoculated with phosphate-solubilizing fungi[J]. Adv Agron, 1999, 69: 99-151. |
| [6] |
Alori ET, Glick BR, Babalola OO. Microbial phosphorus solubilization and its potential for use in sustainable agriculture[J]. Front Microbiol, 2017, 8: 971.
doi: 10.3389/fmicb.2017.00971 pmid: 28626450 |
| [7] |
Xie JG, Yan ZQ, Wang GF, et al. A bacterium isolated from soil in a Karst rocky desertification region has efficient phosphate-solubilizing and plant growth-promoting ability[J]. Front Microbiol, 2021, 11: 625450.
doi: 10.3389/fmicb.2020.625450 URL |
| [8] |
Reyes I, Bernier L, Antoun H. Rock phosphate solubilization and colonization of maize rhizosphere by wild and genetically modified strains of Penicillium rugulosum[J]. Microb Ecol, 2002, 44(1): 39-48.
pmid: 12019460 |
| [9] | 池景良, 郝敏, 王志学, 等. 解磷微生物研究及应用进展[J]. 微生物学杂志, 2021, 41(1): 1-7. |
| Chi JL, Hao M, Wang ZX, et al. Advances in research and application of phosphorus-solubilizing microorganism[J]. J Microbiol, 2021, 41(1): 1-7. | |
| [10] |
王俊娟, 阎爱华, 王薇, 等. 铁尾矿区油松根际溶磷泛菌D2的筛选鉴定及溶磷特性[J]. 应用生态学报, 2016, 27(11): 3705-3711.
doi: 10.13287/j.1001-9332.201611.003 |
| Wang JJ, Yan AH, Wang W, et al. Screening, identification and phosphate-solubilizing characteristics of phosphate-solubilizing bacteria strain D2(Pantoea sp.)in rhizosphere of Pinus tabuliformis in iron tailings yard[J]. Chin J Appl Ecol, 2016, 27(11): 3705-3711. | |
| [11] | 李海云, 姚拓, 张榕, 等. 红三叶根际溶磷菌株分泌有机酸与溶磷能力的相关性研究[J]. 草业学报, 2018, 27(12): 113-121. |
| Li HY, Yao T, Zhang R, et al. Relationship between organic acids secreted from rhizosphere phosphate-solubilizing bacteria in Trifolium pratense and phosphate-solubilizing ability[J]. Acta Prataculturae Sin, 2018, 27(12): 113-121. | |
| [12] | Zhu FL, Qu LY, Hong XG, et al. Isolation and characterization of a phosphate-solubilizing halophilic bacterium Kushneria sp. YCWA18 from daqiao saltern on the coast of Yellow Sea of China[J]. Evid Based Complementary Altern Med, 2011, 2011: 615032. |
| [13] |
Ochoa-Loza FJ, Artiola JF, Maier RM. Stability constants for the complexation of various metals with a rhamnolipid biosurfactant[J]. J Environ Qual, 2001, 30(2): 479-485.
pmid: 11285908 |
| [14] |
Ludueña LM, Anzuay MS, Angelini JG, et al. Strain Serratia sp. S119: a potential biofertilizer for peanut and maize and a model bacterium to study phosphate solubilization mechanisms[J]. Appl Soil Ecol, 2018, 126: 107-112.
doi: 10.1016/j.apsoil.2017.12.024 URL |
| [15] |
Ludueña LM, Anzuay MS, Angelini JG, et al. Role of bacterial pyrroloquinoline quinone in phosphate solubilizing ability and in plant growth promotion on strain Serratia sp. S119[J]. Symbiosis, 2017, 72(1): 31-43.
doi: 10.1007/s13199-016-0434-7 URL |
| [16] |
Chen ML, Ma YK, Wu S, et al. Genome warehouse: a public repository housing genome-scale data[J]. Genomics Proteomics Bioinformatics, 2021, 19(4): 584-589.
doi: 10.1016/j.gpb.2021.04.001 URL |
| [17] | 中华人民共和国农业部. NY/T 2421-2013植株全磷含量测定钼锑抗比色法[S]. 国家标准馆, 2013. |
| Ministry of Agriculture and Rural Affairs of the People's Republic of China. NY/T 2421-2013 Determination of total phosphorus in plant. Vanadium molybdate blue colorimetric method[S]. National Library of Standards, 2013. | |
| [18] | 国家质量监督检验检疫总局, 中国国家标准化管理委员会. 尿素的测定方法第4部分:铁含量邻菲啰啉分光光度法: GB/T 2441.4—2010[S]. 北京: 中国标准出版社, 2011. |
| General Administration of Quality Supervision, Inspection and Quarantine of the People's Republic of China, Standardization Administration of the People's Republic of China. Determination of urea—Part 4: iron content—1, 10-Phenanthroline spectrophotometric method: GB/T 2441.4—2010[S]. Beijing: Standards Press of China, 2011. | |
| [19] | 国家统计局. 第七次全国人口普查公报(第二号)[R]. 2021. |
| National Bureau of Statistics. Communiqué of the Seventh National Population Census(No. 2)[R]. 2021. | |
| [20] |
唐岷宸, 李文静, 宋天顺, 等. 一株高效解磷菌的筛选及其解磷效果验证[J]. 生物技术通报, 2020, 36(6): 102-109.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0969 |
| Tang MC, Li WJ, Song TS, et al. Screening of a highly efficient phosphate-solubilizing bacterium and validation of its phosphate-solubilizing effect[J]. Biotechnol Bull, 2020, 36(6): 102-109. | |
| [21] | 魏烈群. 荣成天鹅湖解磷菌的分离筛选及其对沉积物磷释放的影响[D]. 烟台: 烟台大学, 2021. |
| Wei LQ. Isolation and screening of phosphate-solubilizing bacteria and the effect on phosphorus release from the sediments in Rongcheng Swan lake[D]. Yantai: Yantai University, 2021. | |
| [22] | 朱德旋, 杜春梅, 董锡文, 等. 一株寒地高效解无机磷细菌的分离鉴定及拮抗作用[J]. 微生物学报, 2020, 60(8): 1672-1682. |
| Zhu DX, Du CM, Dong XW, et al. Identification and antagonism activity of an inorganic phosphorus-dissolving bacterial strain isolated from cold region[J]. Acta Microbiol Sin, 2020, 60(8): 1672-1682. | |
| [23] | 赵伟进, 王孝先, 杨洋, 等. 黑青稞根际促生菌筛选及其对种子萌发的影响[J]. 种子, 2018, 37(12): 1-5, 10. |
| Zhao WJ, Wang XX, Yang Y, et al. Selection of rhizotrophic bacteria from rhizosphere and its effect on seed germination of black barley[J]. Seed, 2018, 37(12): 1-5, 10. | |
| [24] |
Zhao JJ, Wang S, Zhu XF, et al. Isolation and characterization of nodules endophytic bacteria Pseudomonas protegens Sneb1997 and Serratia plymuthica Sneb2001 for the biological control of root-knot nematode[J]. Appl Soil Ecol, 2021, 164: 103924.
doi: 10.1016/j.apsoil.2021.103924 URL |
| [25] |
陈佳兴, 秦琴, 邱树毅, 等. 磷尾矿土壤中解磷细菌的筛选及解磷能力的测定[J]. 生物技术通报, 2018, 34(6): 183-189.
doi: 10.13560/j.cnki.biotech.bull.1985.2017-1122 |
| Chen JX, Qin Q, Qiu SY, et al. Isolation, identification of phosphate-solubilizing bacteria derived from phosphate tailing soil and their capacities[J]. Biotechnol Bull, 2018, 34(6): 183-189. | |
| [26] | 卢琴, 罗青平, 张腾飞, 等. 灰雁黏质沙雷菌的鉴定及其耐药性分析[J]. 动物医学进展, 2019, 40(2): 130-134. |
| Lu Q, Luo QP, Zhang TF, et al. Identification and drug sensitivity test of Serratia marcescens in greylag goose[J]. Prog Vet Med, 2019, 40(2): 130-134. | |
| [27] | 乔志伟, 洪坚平, 谢英荷, 等. 石灰性土壤拉恩式溶磷细菌的筛选鉴定及溶磷特性[J]. 应用生态学报, 2013, 24(8): 2294-2300. |
| Qiao ZW, Hong JP, Xie YH, et al. Screening, identification and phosphate-solubilizing characteristics of Rahnella sp. phosphate-solubilizing bacteria in calcareous soil[J]. Chin J Appl Ecol, 2013, 24(8): 2294-2300. |
| [1] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [2] | ZHAO Guang-xu, YANG He-tong, SHAO Xiao-bo, CUI Zhi-hao, LIU Hong-guang, ZHANG Jie. Phosphate-solubilizing Properties and Optimization of Cultivation Conditions of Penicillium rubens: A Highly Efficient Phosphate Solubilizer [J]. Biotechnology Bulletin, 2023, 39(9): 71-83. |
| [3] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [4] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [5] | XIE Dong, WANG Liu-wei, LI Ning-jian, LI Ze-lin, XU Zi-hang, ZHANG Qing-hua. Exploration, Identification and Phosphorus-solubilizing Condition Optimization of a Multifunctional Strain [J]. Biotechnology Bulletin, 2023, 39(7): 241-253. |
| [6] | XU Hong-Yun, LV Jun, YU Cun. Growth Promoting of Pinus massoniana Seedlings Regulated by Rhizosphere Phosphate-solubilizing Paraburkholderia spp. [J]. Biotechnology Bulletin, 2023, 39(6): 274-285. |
| [7] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| [8] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [9] | ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101 [J]. Biotechnology Bulletin, 2023, 39(10): 292-303. |
| [10] | WEN Chang, LIU Chen, LU Shi-yun, XU Zhong-bing, AI Chao-fan, LIAO Han-peng, ZHOU Shun-gui. Biological Characteristics and Genome Analysis of a Novel Multidrug-resistant Shigella flexneri Phage [J]. Biotechnology Bulletin, 2022, 38(9): 127-135. |
| [11] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [12] | SHEN Jia-jia, HOU Xiao-gai, WANG Er-qiang, WANG Fei, GUO Li-li. Organic Phosphate-solubilizing Bacteria Screening in the Rhizosphere of Paeonia ostii and Study on Their Phosphate-solubilizing Capabilities [J]. Biotechnology Bulletin, 2022, 38(6): 157-165. |
| [13] | HU Shan, LIANG Wei-qu, HUANG Hao, XU Cong, LUO Hua-jian, HU Chu-wei, HUANG Xiao-yan, CHEN Shi-li. Screening,Identification and Antagonism of Phosphate-Solubilizing Bacteria from the Compost Chinese Medicinal Herbal Residues [J]. Biotechnology Bulletin, 2022, 38(3): 92-102. |
| [14] | ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9 [J]. Biotechnology Bulletin, 2022, 38(10): 216-225. |
| [15] | CHEN Ti-qiang, XU Xiao-lan, SHI Lin-chun, ZHONG Li-Yi. Sequencing and Analysis of the Whole Genome of Zizhi Cultivar ‘Wuzhi No.2’(Ganoderma sp. strain Zizhi S2) [J]. Biotechnology Bulletin, 2021, 37(11): 42-56. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||