








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (2): 232-242.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0612
Previous Articles Next Articles
HE Meng-ying( ), LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao(
), LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao( )
)
Received:2022-05-19
Online:2023-02-26
Published:2023-03-07
HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43[J]. Biotechnology Bulletin, 2023, 39(2): 232-242.

Fig. 1 Characteristics of strain WA4-43 A: Growth state of strain Gao’s No.1 medium. B: Strain gram staining light microscope(1 000×). C to D: Observed results of 10 000× and 20 000× by scanning electron microscope
| 检测项目 Test item | 结果 Result | 检测项目 Test item | 结果 Result | |
|---|---|---|---|---|
| 明胶水解 | - | 氧化酶 | - | |
| 鼠李糖 | + | 过氧化氢酶 | + | |
| 葡萄糖 | + | 尿素酶 | + | |
| 木糖 | + | 生长耐受NaCl范围 | 1%-5% | |
| 甘露醇 | + | H2S产生 | - |
Table 1 Physiological and biochemical characteristics of strain WA4-43
| 检测项目 Test item | 结果 Result | 检测项目 Test item | 结果 Result | |
|---|---|---|---|---|
| 明胶水解 | - | 氧化酶 | - | |
| 鼠李糖 | + | 过氧化氢酶 | + | |
| 葡萄糖 | + | 尿素酶 | + | |
| 木糖 | + | 生长耐受NaCl范围 | 1%-5% | |
| 甘露醇 | + | H2S产生 | - |

Fig. 3 Determination of antibacterial activity of strain WA4-43 by tube and plate method A: Methicillin-resistant Staphylococcus aureus. B: Staphylococcus aureus. C: Bacillus subtilis. D: Staphylococcu epidermidis. n=3
| 数据库 Database | 数量 Amount | 100≤长度<300 100≤Length <300 | 长度≥300 Length≥300 |
|---|---|---|---|
| eggnog Annotation | 4 178 | 1 847 | 2 181 |
| GO Annotation | 3 563 | 1 539 | 1 892 |
| Kegg Annotation | 1 870 | 686 | 1 117 |
| Nr Annotation | 4 835 | 2 230 | 2 311 |
| Pfam Annotation | 3 239 | 1 398 | 1 735 |
| Swissprot Annotation | 2 591 | 954 | 1 570 |
| TrEMBL Annotation | 2 591 | 954 | 1 570 |
| All Annotated | 4 837 | 2 230 | 2 311 |
Table 2 Number and size of functionally annotated genes in strain WA4-43
| 数据库 Database | 数量 Amount | 100≤长度<300 100≤Length <300 | 长度≥300 Length≥300 |
|---|---|---|---|
| eggnog Annotation | 4 178 | 1 847 | 2 181 |
| GO Annotation | 3 563 | 1 539 | 1 892 |
| Kegg Annotation | 1 870 | 686 | 1 117 |
| Nr Annotation | 4 835 | 2 230 | 2 311 |
| Pfam Annotation | 3 239 | 1 398 | 1 735 |
| Swissprot Annotation | 2 591 | 954 | 1 570 |
| TrEMBL Annotation | 2 591 | 954 | 1 570 |
| All Annotated | 4 837 | 2 230 | 2 311 |
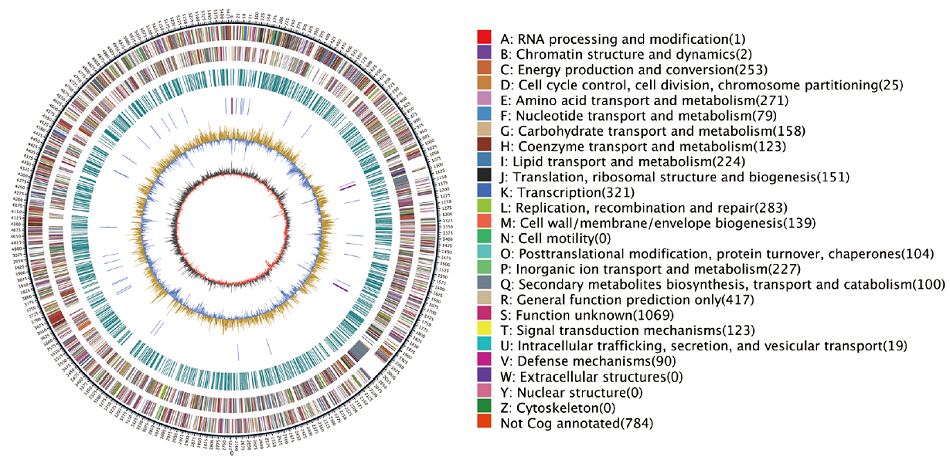
Fig. 4 Genome circle map of strain WA4-43 Outer circle: Genome size. The second circle: Size of all plus-strand genes in the genome.The third circle: The size of all negative-strand genes in the genome.The fourth circle: Genome sequence repeats.The fifth circle: 49 blue TrNAand purple RRNA.The sixth circle: GC content. Innermost circle: GC-SKEw
| 基因簇 Region | 类型 Type | 预测产生的化合物 Predicited production | 同源性 Similarity/% |
|---|---|---|---|
| BGC1 | Ectoine | Ectoine | 75 |
| BGC2 | Terpene | SF2575 | 6 |
| BGC3 | Arylpolyene | ||
| BGC4 | NRPS, betalactone | ||
| BGC5 | NRPS | Ishigamide | 11 |
| BGC6 | redox-cofactor | ||
| BGC7 | NRPS | ||
| BGC8 | NAPAA | ||
| BGC9 | Terpene | Oxalomycin B | 6 |
| BGC10 | RiPP-like | Kanglemycin A/Kanglemycin V1/Kanglemycin V2 | 5 |
| BGC11 | T1PKS, NRPS-like | ||
| BGC12 | NRPS, siderophore | Desferrioxamine | 33 |
| BGC13 | NRPS |
Table 3 Prediction of secondary metabolic gene clusters in the genome of strain WA4-43
| 基因簇 Region | 类型 Type | 预测产生的化合物 Predicited production | 同源性 Similarity/% |
|---|---|---|---|
| BGC1 | Ectoine | Ectoine | 75 |
| BGC2 | Terpene | SF2575 | 6 |
| BGC3 | Arylpolyene | ||
| BGC4 | NRPS, betalactone | ||
| BGC5 | NRPS | Ishigamide | 11 |
| BGC6 | redox-cofactor | ||
| BGC7 | NRPS | ||
| BGC8 | NAPAA | ||
| BGC9 | Terpene | Oxalomycin B | 6 |
| BGC10 | RiPP-like | Kanglemycin A/Kanglemycin V1/Kanglemycin V2 | 5 |
| BGC11 | T1PKS, NRPS-like | ||
| BGC12 | NRPS, siderophore | Desferrioxamine | 33 |
| BGC13 | NRPS |
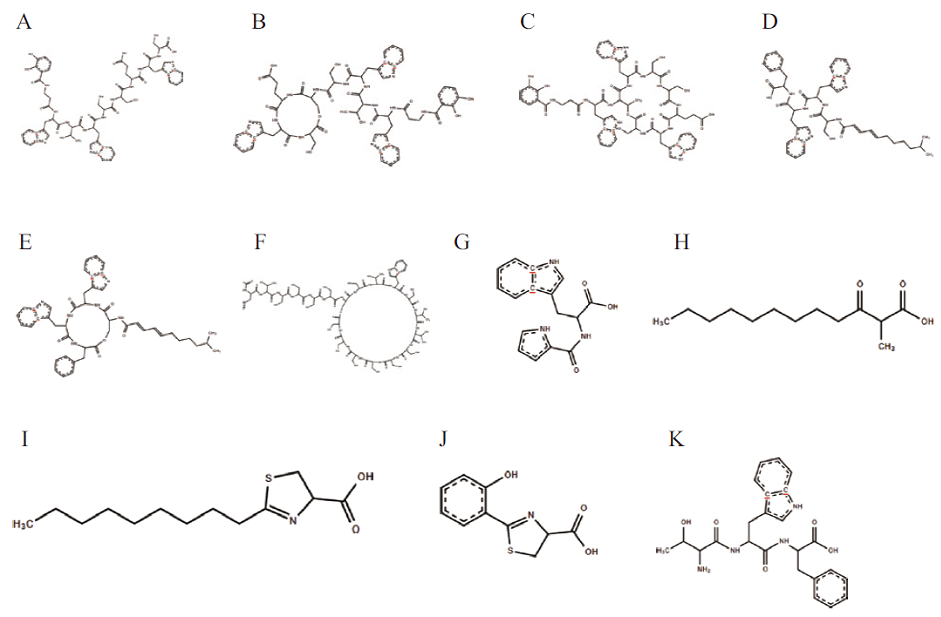
Fig. 8 Prediction of biosynthetic gene cluster compounds of strain WA4-43 A to C: Predict gene cluster 4 compound. D to E: Predict gene cluster 5 compound. F: Predict gene cluster 6 compound. G: Predict gene cluster 7 compound. H-J: Predict gene cluster 11 compound. K: Predict gene cluster 12 compound
| 基因编号 Gene ID | 基因功能 Gene function | 长度 Length/bp | Nr数据库 Nr database | GO数据库 GO database | TrEMBL数据库 TrEMBL database |
|---|---|---|---|---|---|
| GE002623 | Additional biosynthetic genes | 1 479 | Gordonia sp. IITR100 | Transferase activity;carbon-nitrogen ligase activity, with glutamine as amido-N-donor | Amidase |
| GE002624 | Other genes | 1 674 | Gordonia sp. UCD-TK1 | oxidoreductase activity, acting on the CH-CH group of donors; oxidation-reduction process | FAD-binding dehydrogenase |
| GE002625 | Other genes | 1 164 | Gordonia sp. v-85 | peroxidase activity; heme binding; oxidation-reduction process; cellular oxidant detoxification | Putative Iron-dependent peroxidase |
| GE002626 | Core biosynthetic genes | 810 | Nocardia farcinica | proteolysis; peptidase activity; defense response to bacterium | Bacteriocin |
| GE002627 | Transport-related genes | 1 734 | Nocardia farcinica | ATP binding; integral component of membrane; ATPase activity, coupled to transmembrane movement of substances; transmembrane transport | Multidrug ABC transporter ATP-binding protein |
| GE002628 | Transport-related genes | 1 920 | Gordonia sp. IITR100 | ATP binding; integral component of membrane; ATPase activity, coupled to transmembrane movement of substances;transmembrane transport | ATP-binding cassette, subfamily B |
| GE002629 | Other genes | 573 | Gordonia sp. UCD-TK1 | Uncharacterized protein |
Table 4 BGC10 gene prediction and database analysis
| 基因编号 Gene ID | 基因功能 Gene function | 长度 Length/bp | Nr数据库 Nr database | GO数据库 GO database | TrEMBL数据库 TrEMBL database |
|---|---|---|---|---|---|
| GE002623 | Additional biosynthetic genes | 1 479 | Gordonia sp. IITR100 | Transferase activity;carbon-nitrogen ligase activity, with glutamine as amido-N-donor | Amidase |
| GE002624 | Other genes | 1 674 | Gordonia sp. UCD-TK1 | oxidoreductase activity, acting on the CH-CH group of donors; oxidation-reduction process | FAD-binding dehydrogenase |
| GE002625 | Other genes | 1 164 | Gordonia sp. v-85 | peroxidase activity; heme binding; oxidation-reduction process; cellular oxidant detoxification | Putative Iron-dependent peroxidase |
| GE002626 | Core biosynthetic genes | 810 | Nocardia farcinica | proteolysis; peptidase activity; defense response to bacterium | Bacteriocin |
| GE002627 | Transport-related genes | 1 734 | Nocardia farcinica | ATP binding; integral component of membrane; ATPase activity, coupled to transmembrane movement of substances; transmembrane transport | Multidrug ABC transporter ATP-binding protein |
| GE002628 | Transport-related genes | 1 920 | Gordonia sp. IITR100 | ATP binding; integral component of membrane; ATPase activity, coupled to transmembrane movement of substances;transmembrane transport | ATP-binding cassette, subfamily B |
| GE002629 | Other genes | 573 | Gordonia sp. UCD-TK1 | Uncharacterized protein |
| 菌株 Strain | 序号 No. | 基因组大小 Size/Mb | GC含量 GC/% | 染色体 Contigs | rRNA数量 rRNA genes | tRNA数量 tRNA genes | 编码蛋白区数量 Number of coding sequence |
|---|---|---|---|---|---|---|---|
| WA4-43 | GCA 020520305.1 | 5.44 | 67.76 | 1 | 9 | 49 | 4 963 |
| NCTC10669 | GCA 901542405.1 | 5.71 | 67.8 | 2 | 12 | 46 | 4 974 |
| NRRL B-16283 | GCA 003183825.1 | 5.71 | 67.8 | 1 | 9 | 46 | 4 979 |
| 3612 | GCA 001698225.1 | 5.70 | 67.8 | 1 | 9 | 46 | 4 977 |
| RL-JC02 | GCA 011290605.1 | 5.31 | 67.87 | 2 | 9 | 47 | 4 627 |
| NRRL B-16283 | GCA 000716975.1 | 5.67 | 67.8 | 108 | 7 | 46 | 4 938 |
| UMB0777 | GCA 002847865.1 | 5.70 | 67.7 | 180 | 3 | 46 | 5 007 |
| K | GCA 005502725.1 | 5.19 | 68 | 155 | 9 | 45 | 4 480 |
| C-6 | GCA 000390025.1 | 5.17 | 67.9 | 130 | 5 | 45 | 4 572 |
| NBRC 100016 | GCA 000248035.2 | 5.67 | 67.8 | 130 | 6 | 46 | 4 956 |
Table 5 Comparison of genomic characteristics of 10 Gordonia terrae species
| 菌株 Strain | 序号 No. | 基因组大小 Size/Mb | GC含量 GC/% | 染色体 Contigs | rRNA数量 rRNA genes | tRNA数量 tRNA genes | 编码蛋白区数量 Number of coding sequence |
|---|---|---|---|---|---|---|---|
| WA4-43 | GCA 020520305.1 | 5.44 | 67.76 | 1 | 9 | 49 | 4 963 |
| NCTC10669 | GCA 901542405.1 | 5.71 | 67.8 | 2 | 12 | 46 | 4 974 |
| NRRL B-16283 | GCA 003183825.1 | 5.71 | 67.8 | 1 | 9 | 46 | 4 979 |
| 3612 | GCA 001698225.1 | 5.70 | 67.8 | 1 | 9 | 46 | 4 977 |
| RL-JC02 | GCA 011290605.1 | 5.31 | 67.87 | 2 | 9 | 47 | 4 627 |
| NRRL B-16283 | GCA 000716975.1 | 5.67 | 67.8 | 108 | 7 | 46 | 4 938 |
| UMB0777 | GCA 002847865.1 | 5.70 | 67.7 | 180 | 3 | 46 | 5 007 |
| K | GCA 005502725.1 | 5.19 | 68 | 155 | 9 | 45 | 4 480 |
| C-6 | GCA 000390025.1 | 5.17 | 67.9 | 130 | 5 | 45 | 4 572 |
| NBRC 100016 | GCA 000248035.2 | 5.67 | 67.8 | 130 | 6 | 46 | 4 956 |
| [1] |
Chevrette MG, Carlson CM, Ortega HE, et al. The antimicrobial potential of Streptomyces from insect microbiomes[J]. Nat Commun, 2019, 10(1): 516.
doi: 10.1038/s41467-019-08438-0 pmid: 30705269 |
| [2] |
Jose PA, Jebakumar SRD. Non-streptomycete actinomycetes nourish the current microbial antibiotic drug discovery[J]. Front Microbiol, 2013, 4: 240.
doi: 10.3389/fmicb.2013.00240 pmid: 23970883 |
| [3] |
Hu DN, Gao C, Sun CH, et al. Genome-guided and mass spectrometry investigation of natural products produced by a potential new actinobacterial strain isolated from a mangrove ecosystem in Futian, Shenzhen, China[J]. Sci Rep, 2019, 9(1): 823.
doi: 10.1038/s41598-018-37475-w pmid: 30696899 |
| [4] |
Xiao YS, Zhang B, Zhang M, et al. Rifamorpholines A-E, potential antibiotics from locust-associated actinobacteria Amycolatopsis sp. Hca4[J]. Org Biomol Chem, 2017, 15(18): 3909-3916.
doi: 10.1039/C7OB00614D URL |
| [5] |
Ishii S. Ecology of pathogens and antibiotic-resistant bacteria in environments: challenges and opportunities[J]. Microbes Environ, 2019, 34(1): 1-4.
doi: 10.1264/jsme2.ME3401rh pmid: 30930405 |
| [6] | Kwon SJ, Choi YJ, Kim JM, et al. Complete genome sequence of the carotenoid-producing strain Gordonia ajoucoccus A2[J]. Microbiol Resour Announc, 2020, 9(37): e00662-e00620. |
| [7] |
Chen SQ, Zhao CC, Liu QY, et al. Biodesulfurization of diesel oil in oil-water two phase reaction system by Gordonia sp. SC-10[J]. Biotechnol Lett, 2019, 41(4/5): 547-554.
doi: 10.1007/s10529-019-02663-9 URL |
| [8] | 周东琴, 魏德洲. 沟戈登氏菌对重金属的生物吸附-浮选和解吸性能[J]. 环境科学, 2006, 27(5): 960-964. |
| Zhou DQ, Wei DZ. Biosorptive-flotation and desorption operation of heavy metals from wastewater effluents by gordona amarae[J]. Environ Sci, 2006, 27(5): 960-964. | |
| [9] | 刘凌燕. 美洲大蠊肠道内生戈登氏放线菌WA8-44次级代谢产物抗真菌活性的初步研究[D]. 广州: 广东药科大学, 2019. |
| Liu LY. Preliminary study on the antifungal activity of the secondary metabolites of Gordonia WA8-44 from the intestinal tract of Periplaneta americana[D]. Guangzhou: Guangdong Pharmaceutical University, 2019. | |
| [10] |
Ma Y, Xu MH, Liu HC, et al. Antimicrobial compounds were isolated from the secondary metabolites of Gordonia, a resident of intestinal tract of Periplaneta americana[J]. AMB Express, 2021, 11(1): 111.
doi: 10.1186/s13568-021-01272-y URL |
| [11] |
Challis GL. Exploitation of the Streptomyces coelicolor A3(2)genome sequence for discovery of new natural products and biosynthetic pathways[J]. J Ind Microbiol Biotechnol, 2014, 41(2): 219-232.
doi: 10.1007/s10295-013-1383-2 URL |
| [12] | Becerril A, Álvarez S, Braña AF, et al. Uncovering production of specialized metabolites by Streptomyces argillaceus: activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches[J]. PLoS One, 2018, 13(5): e0198145. |
| [13] | 刘洋. 海洋链霉菌Streptomyces Sp. B59中天然产物的挖掘[D]. 济南: 山东大学, 2019. |
| Liu Y. Mining of natural products from A marine Streptomyces Sp. B59[D]. Jinan: Shandong University, 2019. | |
| [14] |
Karita S, Nakayama K, Goto M, et al. A novel cellulolytic, anaerobic, and thermophilic bacterium, Moorella sp. strain F21[J]. Biosci Biotechnol Biochem, 2003, 67(1): 183-185.
doi: 10.1271/bbb.67.183 URL |
| [15] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [16] | 焦世耀, 张兰威, 李春. 管碟法测定nisin效价的改进[J]. 食品科学, 2005, 26(7): 175-176. |
| Jiao SY, Zhang LW, Li C. Improved Oxford assay on estimation of nisin[J]. Food Sci, 2005, 26(7): 175-176. | |
| [17] |
Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation[J]. Genome Res, 2017, 27(5): 722-736.
doi: 10.1101/gr.215087.116 URL |
| [18] |
Kalvari I, Argasinska J, Quinones-Olvera N, et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families[J]. Nucleic Acids Res, 2018, 46(D1): D335-D342.
doi: 10.1093/nar/gkx1038 URL |
| [19] |
Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes[J]. Nucleic Acids Res, 2016, 44(W1): W54-W57.
doi: 10.1093/nar/gkw413 URL |
| [20] |
Fu LM, Niu BF, Zhu ZW, et al. CD-HIT: accelerated for clustering the next-generation sequencing data[J]. Bioinformatics, 2012, 28(23): 3150-3152.
doi: 10.1093/bioinformatics/bts565 pmid: 23060610 |
| [21] |
Conesa A, Götz S, García-Gómez JM, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research[J]. Bioinformatics, 2005, 21(18): 3674-3676.
doi: 10.1093/bioinformatics/bti610 pmid: 16081474 |
| [22] |
Powell S, Forslund K, Szklarczyk D, et al. eggNOG v4.0: nested orthology inference across 3686 organisms[J]. Nucleic Acids Res, 2014, 42(Database issue): D231-D239.
doi: 10.1093/nar/gkt1253 URL |
| [23] |
Mistry J, Chuguransky S, Williams L, et al. Pfam: The protein families database in 2021[J]. Nucleic Acids Res, 2021, 49(D1): D412-D419.
doi: 10.1093/nar/gkaa913 pmid: 33125078 |
| [24] | Yang MZ, Derbyshire MK, Yamashita RA, et al. NCBI’s conserved domain database and tools for protein domain analysis[J]. Curr Protoc Bioinformatics, 2020, 69(1): e90. |
| [25] |
Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome[J]. Nucleic Acids Res, 2004, 32(Datab-ase issue): D277-D280.
doi: 10.1093/nar/gkh063 URL |
| [26] |
Hyatt D, Chen GL, Locascio PF, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification[J]. BMC Bioinformatics, 2010, 11: 119.
doi: 10.1186/1471-2105-11-119 pmid: 20211023 |
| [27] | Lee N, Hwang S, Kim J, et al. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces[J]. Comput Struct Biotechnol J, 2020, 18: 1548-1556. |
| [28] |
Skinnider MA, Johnston CW, Gunabalasingam M, et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences[J]. Nat Commun, 2020, 11(1): 6058.
doi: 10.1038/s41467-020-19986-1 pmid: 33247171 |
| [29] | Boone DR, Castenholz RW, Garrity GM. Bergey’s Manual® of Systematic Bacteriology[M]. 2rd ed.ed. New York: Springer, 2001. |
| [30] |
Zhang HY, Lin Z, Liu B, et al. Bioremediation of di-(2-ethylhexyl)phthalate contaminated red soil by Gordonia terrae RL-JC02: Characterization, metabolic pathway and kinetics[J]. Sci Total Environ, 2020, 733: 139138.
doi: 10.1016/j.scitotenv.2020.139138 URL |
| [31] | 曹然, 罗晓霞, 张利莉. 链霉菌新物种Streptomyces wensuensis TRM68367全基因组测序及生物信息学分析[J]. 塔里木大学学报, 2021, 33(3): 1-9. |
| Cao R, Luo XX, Zhang LL. Whole genome sequencing and bioinformatics analysis of a novel actinomycete Streptomyces wensuensis TRM68367[J]. J Tarim Univ, 2021, 33(3): 1-9. | |
| [32] | 张鑫, 舒志万, 李永臻, 等. 相容溶质四氢嘧啶的微生物合成研究进展[J]. 生物工程学报, 2022, 38(3): 868-881. |
| Zhang X, Shu ZW, Li YZ, et al. Advances in the microbial production of the compatible solute ectoine: a review[J]. Chin J Biotechnol, 2022, 38(3): 868-881. | |
| [33] | Li L, Wang P, Tang Y. C-glycosylation of anhydrotetracycline scaffold with SsfS6 from the SF2575 biosynthetic pathway[J]. J Antibiot(Tokyo), 2014, 67(1): 65-70. |
| [34] |
Zhao CH, Ju JH, Christenson SD, et al. Utilization of the methoxymalonyl-acyl carrier protein biosynthesis locus for cloning the oxazolomycin biosynthetic gene cluster from Streptomyces albus JA3453[J]. J Bacteriol, 2006, 188(11): 4142-4147.
doi: 10.1128/JB.00173-06 URL |
| [35] | Becerril A, Álvarez S, Braña AF, et al. Uncovering production of specialized metabolites by Streptomyces argillaceus: Activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches[J]. PLoS One, 2018, 13(5): e0198145. |
| [36] | Harbottle J, Mosaei H, Allenby N, et al. Kanglemycin A can overcome rifamycin resistance caused by ADP-ribosylation by arr protein[J]. Antimicrob Agents Chemother, 2021, 65(12): e0086421. |
| [37] |
Aparicio JF, Fouces R, et al. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis[J]. Chem Biol, 2000, 7(11): 895-905.
doi: 10.1016/S1074-5521(00)00038-7 URL |
| [38] | Kloosterman AM, Shelton KE, van Wezel GP, et al. RRE-finder: a genome-mining tool for class-independent RiPP discovery[J]. mSystems, 2020, 5(5): e00267-e00220. |
| [1] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [2] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [3] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [4] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| [5] | ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101 [J]. Biotechnology Bulletin, 2023, 39(10): 292-303. |
| [6] | WANG Shuai, LV Hong-rui, ZHANG Hao, WU Zhan-wen, XIAO Cui-hong, SUN Dong-mei. Whole-Genome Sequencing Identification of Phosphate-solubilizing Bacteria PSB-R and Analysis of Its Phosphate-solubilizing Properties [J]. Biotechnology Bulletin, 2023, 39(1): 274-283. |
| [7] | WEN Chang, LIU Chen, LU Shi-yun, XU Zhong-bing, AI Chao-fan, LIAO Han-peng, ZHOU Shun-gui. Biological Characteristics and Genome Analysis of a Novel Multidrug-resistant Shigella flexneri Phage [J]. Biotechnology Bulletin, 2022, 38(9): 127-135. |
| [8] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [9] | ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9 [J]. Biotechnology Bulletin, 2022, 38(10): 216-225. |
| [10] | WANG Nan, SU Yu, LIU Wen-jie, FENG Ming, MAO Yu, ZHANG Xin-guo. Research Progress on Active Compounds Against Drug-resistant Microorganism from Plant Endophytes [J]. Biotechnology Bulletin, 2021, 37(8): 263-274. |
| [11] | LIANG Zhen-ting, TANG Ting. Effects of Endophytes on Biosynthesis of Secondary Metabolites and Stress Tolerance in Plants [J]. Biotechnology Bulletin, 2021, 37(8): 35-45. |
| [12] | CHEN Ti-qiang, XU Xiao-lan, SHI Lin-chun, ZHONG Li-Yi. Sequencing and Analysis of the Whole Genome of Zizhi Cultivar ‘Wuzhi No.2’(Ganoderma sp. strain Zizhi S2) [J]. Biotechnology Bulletin, 2021, 37(11): 42-56. |
| [13] | GUO He-bao, WANG Xing, HE Shan-wen, ZHANG Xiao-xia. Phenotypic Characteristics Combined with Genomic Analysis to Identify Different Colony Morphology Bacillus velezensis ACCC 19742 [J]. Biotechnology Bulletin, 2020, 36(2): 142-148. |
| [14] | ZHAO Jiang-hua, FANG Huan, ZHANG Da-wei. Research Progress in Biosynthesis of Secondary Metabolites of Microorganisms [J]. Biotechnology Bulletin, 2020, 36(11): 141-147. |
| [15] | WANG Ya-li, ZHU Shan-shan, YANG Feng-shan, MA Yu-kun, FU Hai-yan, LIU Chun-guang. Pan-genome Sequencing and Comparative Genomic Analysis of Atrazine-degrading Bacteria [J]. Biotechnology Bulletin, 2019, 35(7): 90-99. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||