








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (8): 85-94.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1386
Previous Articles Next Articles
CEN You-fei1,2( ), ZHU Mu-zi2, YE Wei2, LI Sai-ni2, ZHONG Guo-hua1(
), ZHU Mu-zi2, YE Wei2, LI Sai-ni2, ZHONG Guo-hua1( ), ZHANG Wei-min2(
), ZHANG Wei-min2( )
)
Received:2020-11-14
Online:2021-08-26
Published:2021-09-10
Contact:
ZHONG Guo-hua,ZHANG Wei-min
E-mail:297288514@qq.com;guohuazhong@scau.edu.cn;wmzhang@gdim.cn
CEN You-fei, ZHU Mu-zi, YE Wei, LI Sai-ni, ZHONG Guo-hua, ZHANG Wei-min. Cloning and Functional Identification of Trichothecene Mycotoxin Biosynthesis Gene Promoter from Paramyrothecium roridum[J]. Biotechnology Bulletin, 2021, 37(8): 85-94.
| 目的基因Target gene | 反向引物Reverse primer sp1(5'-3') | 反向引物Reverse primer sp2(5'-3') | 反向引物Reverse primer sp3(5'-3') |
|---|---|---|---|
| tri5 | TGTACTTGACGTTCTCCAGCAAC | GGGAGATTCTGAGAGAGATAGATA | TCTGTCTGTCTGATCTGTCTGTC |
| tri12 | GCGTTGGTCAACATGAAGAATGG | ACATCTGCGGCGATGTTTGAGAT | ATCAAACGAGGGCTCCGATAGTA |
Table 1 Specific reverse primer sequences of the target genes tri5 and tri12
| 目的基因Target gene | 反向引物Reverse primer sp1(5'-3') | 反向引物Reverse primer sp2(5'-3') | 反向引物Reverse primer sp3(5'-3') |
|---|---|---|---|
| tri5 | TGTACTTGACGTTCTCCAGCAAC | GGGAGATTCTGAGAGAGATAGATA | TCTGTCTGTCTGATCTGTCTGTC |
| tri12 | GCGTTGGTCAACATGAAGAATGG | ACATCTGCGGCGATGTTTGAGAT | ATCAAACGAGGGCTCCGATAGTA |
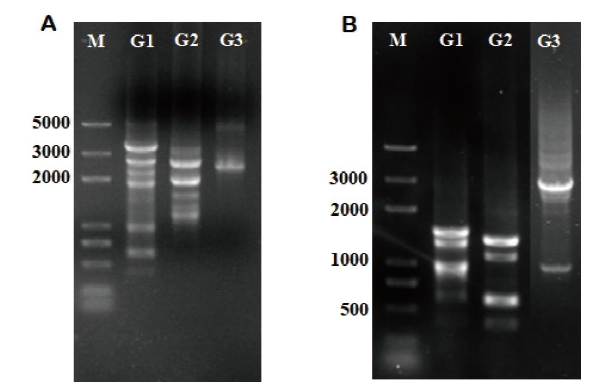
Fig.1 Target fragments of tri5 and tri12 obtained by chromosome walking M:Marker trans2K plus;A:The PCR amplification products of tri5 after three rounds of chromosome walking;B:The PCR amplification products of tri12 after three rounds of chromosome walking
| 目的基因启动子 Promoter of target gene | 分值 Score | 核心序列 Core sequence |
|---|---|---|
| tri5 | 0.71 | ACCAGAGACGGGAATCACGCGCATT- TCTGGCTACGATATCCCCCCTAACG |
| tri12 | 0.98 | AGGCCCTCGATAAGAAGCGGCGGCG- GGGCCTCGACCAGGAATCGACTACC |
Table 2 Core sequences of tri5 and tri12 promoters
| 目的基因启动子 Promoter of target gene | 分值 Score | 核心序列 Core sequence |
|---|---|---|
| tri5 | 0.71 | ACCAGAGACGGGAATCACGCGCATT- TCTGGCTACGATATCCCCCCTAACG |
| tri12 | 0.98 | AGGCCCTCGATAAGAAGCGGCGGCG- GGGCCTCGACCAGGAATCGACTACC |
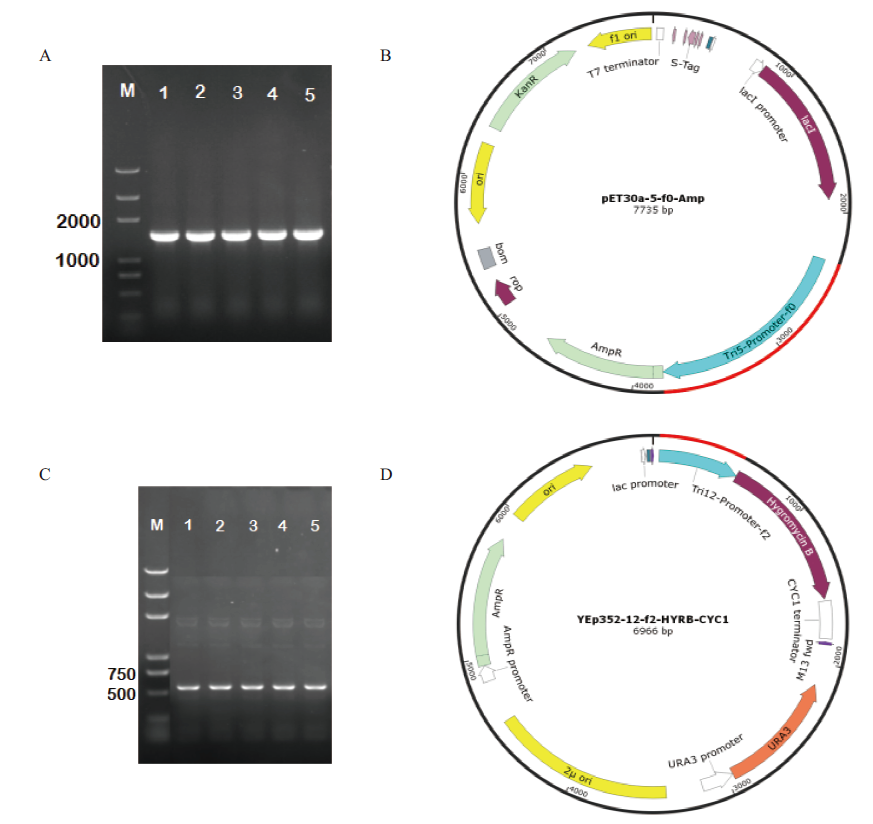
Fig.3 Colony PCR verification of DH5α and promoter vector maps M:Marker trans2K plus. A:Colony PCR of DH5α-pET30a-5-f0-Amp. B:Vector map of pET30a-5-f0-Amp. C:Colony PCR of DH5α-YEp352-12-f2-HYRB-CYC1. D:Vector map of YEp352-12-f2-HYRB-CYC1

Fig.4 Screening of E. coli containing different promoter fragments on resistance plates with different concentrations of ampicillin A:The result of culture on plates of DH5α-pET30a-5-f0/5-f1/5-f2/12-f0/12-f1/12-f2-Amp. B:The result of culture on plates of DH5α-pET30a-5-f0/5-f1/5-f2-Amp. C:OD value bar graph of DH5α-YEp352-12-f1-HYRB-CYC1 and DH5α-pET30a-12-f1-Amp. N:DH5α-pET30a-ACP empty carrier(Negative control);P:DH5α-YEp352-12-f1-HYRB-CYC1(Positive control);5(f0-f2):DH5α-pET30a-5-f0/5-f1/ 5-f2-Amp;12(f0-f2):DH5α-pET30a-12-f0/12-f1/12-f2-Amp
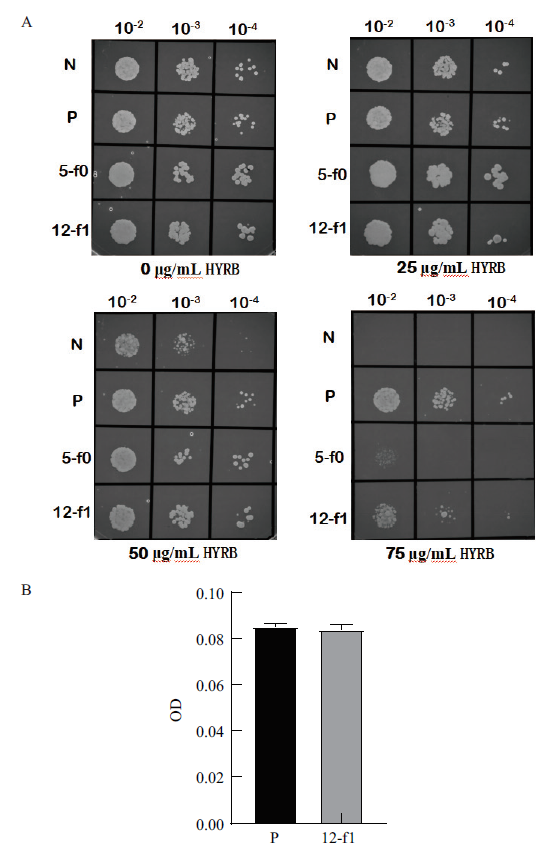
Fig.5 Screening of S. cerevisiae containing different prom-oter fragments on resistant plates with different concentrations of hygromycin A:The result of culture on plates of BJ5464-YEp352-TEF1-HYRB-CYC1(Positive control),BJ5464-YEp352-No pro-HYRB-CYC1(Negative control),BJ5464-YEp352-12-f1-HYRB-CYC1 and BJ5464-YEp352-5-f0-HYRB-CYC1;B:OD value bar graph of BJ5464-YEp352-TEF1-HYRB-CYC1(Positive control)and BJ5464-YEp352-12-f1-HYRB-CYC1. N:BJ5464-YEp352-No pro-HYRB-CYC1;P:BJ5464-YEp352-TEF1-HYRB-CYC1;5-f0:BJ5464-YEp352-5-f0-HYRB-CYC1;12-f1:BJ5464-YEp352-12-f1-HYRB-CYC1
| [1] | Brown DW, Villani A, Susca A, et al. Gain and loss of a transcription factor that regulates late trichothecene biosynthetic pathway genes in Fusarium[J]. Fungal Genetics and Biology, 2020, 136:553-560. |
| [2] |
Cardoza RE, McCormick SP, Lindo L, et al. A cytochrome P450 monooxygenase gene required for biosynjournal of the trichothecene toxin harzianum A in Trichoderma[J]. Applied Microbiology and Biotechnology, 2019, 103(19):8087-8103.
doi: 10.1007/s00253-019-10047-2 pmid: 31384992 |
| [3] |
Lee SR, Seok S, Ryoo R, et al. Macrocyclic trichothecene mycotoxins from a deadly poisonous mushroom, Podostroma cornu-damae[J]. Journal of Natural Products, 2019, 82(1):122-128.
doi: 10.1021/acs.jnatprod.8b00823 URL |
| [4] |
Nguyen LTT, Jang JY, Kim TY, et al. Nematicidal activity of verrucarin A and roridin A isolated from Myrothecium verrucaria against Meloidogyne incognita[J]. Pesticide Biochemistry and Physiology, 2018, 148:133-143.
doi: S0048-3575(17)30508-4 pmid: 29891364 |
| [5] |
Liu HX, Liu WZ, Chen YC, et al. Cytotoxic trichothecene macrolides from the endophyte fungus Myrothecium roridum[J]. Journal of Asian Natural Products Research, 2016, 18(7):684-692.
doi: 10.1080/10286020.2015.1134505 URL |
| [6] | Ye W, Chen YC, Li HH, et al. Two trichothecene mycotoxins from Myrothecium roridum induce apoptosis of HepG-2 cells via caspase activation and disruption of mitochondrial membrane potential[J]. Molecules, 2016, 21(6):E781. |
| [7] |
Cane DE. Enzymic formation of sesquiterpenes[J]. Chemical Reviews, 1990, 90(7):1089-1103.
doi: 10.1021/cr00105a002 URL |
| [8] |
McCormick SP, Alexander NJ, Trapp SE, et al. Disruption of TRI101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides[J]. Applied and Environmental Microbiology, 1999, 65(12):5252-5256.
pmid: 10583973 |
| [9] |
Proctor RH, McCormick SP, Kim HS, et al. Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi[J]. PLoS Pathogens, 2018, 14(4):e1006946.
doi: 10.1371/journal.ppat.1006946 URL |
| [10] |
Trassaert M, Vandermies M, Carly F, et al. New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica[J]. Microbial Cell Factories, 2017, 16(1):141.
doi: 10.1186/s12934-017-0755-0 pmid: 28810867 |
| [11] |
Cooper SJ, Trinklein ND, Anton ED, et al. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome[J]. Genome Research, 2006, 16(1):1-10.
pmid: 16344566 |
| [12] |
Even DY, Kedmi A, Ideses D, et al. Functional screening of core promoter activity[J]. Methods in Molecular Biology, 2017, 1651:77-91.
doi: 10.1007/978-1-4939-7223-4_7 pmid: 28801901 |
| [13] |
Balabanova LA, Shkryl YN, Slepchenko LV, et al. Development of host strains and vector system for an efficient genetic transformation of filamentous fungi[J]. Plasmid, 2019, 101:1-9.
doi: S0147-619X(18)30089-1 pmid: 30465791 |
| [14] | 朱利, 王义, 殷金瑶, 等. 橡胶树白粉菌内源强启动子WY193的克隆及功能鉴定[J]. 分子植物育种, 2020, 18(9):2865-2871. |
| Zhu L, Wang Y, Yin JY, et al. Cloning and function identification of endogenous strong promoter WY193 of Oidium heveae steinm[J]. Molecular Plant Breeding, 2020, 18(9):2865-2871. | |
| [15] | 阎隆飞, 张玉麟. 分子生物学[M]. 第2 版. 北京: 中国农业大学出版社, 1997. |
| Yan LF, Zhang YL. Molecular biology[M]. 2rd ed. Beijing: China Agricultural University Press, 1997. | |
| [16] | Ye W, Liu TM, Zhu MZ, et al. De Novo transcriptome analysis of plant pathogenic fungus Myrothecium roridum and identification of genes associated with trichothecene mycotoxin biosynjournal[J]. International Journal of Molecular Sciences, 2017, 18(3):E497. |
| [17] | Ashrafi M, Fardsi M, Mirshamsi A, et al. Isolation and sequence analysis of gpdII promoter of the white button mushroom(Agaricus bisporus)from strains Holland737 and IM008[J]. International Journal of Horticultural Science and Technology, 2015, 2(1):33-41. |
| [18] |
Madhavan A, Pandey A, et al. Expression system for heterologous protein expression in the filamentous fungus Aspergillus unguis[J]. Bioresource Technology, 2017, 245:1334-1342.
doi: S0960-8524(17)30808-8 pmid: 28578805 |
| [19] |
Keränen S, Penttilä M. Production of recombinant proteins in the filamentous fungus Trichoderma reesei[J]. Current Opinion in Biotechnology, 1995, 6(5):534-537.
pmid: 7579664 |
| [20] | 黄自磊, 章卫民, 叶伟, 等. 深海真菌Dichotomomyces cejpii胶霉毒素生物合成基因启动子的克隆和功能鉴定[J]. 生物技术通报, 2018, 34(4):144-150. |
| Huang ZL, Zhang WM, Ye W, et al. Cloning and identification of gene promoter for gliotoxin biosynjournal from deep-sea-derived fungus Dichotomomyces cejpii[J]. Biotechnology Bulletin, 2018, 34(4):144-150. |
| [1] | ZHOU Lu-qi, CUI Ting-ru, HAO Nan, ZHAO Yu-wei, ZHAO Bin, LIU Ying-chao. Application of Chemical Proteomics in Identifying the Molecular Targets of Natural Products [J]. Biotechnology Bulletin, 2023, 39(9): 12-26. |
| [2] | LIU Yu-ling, WANG Meng-yao, SUN Qi, MA Li-hua, ZHU Xin-xia. Effect of RD29A Promoter on the Stress Resistance of Transgenic Tobacco with SikCDPK1 Gene from Saussurea involucrata [J]. Biotechnology Bulletin, 2023, 39(9): 168-175. |
| [3] | JIANG Hai-rong, CUI Ruo-qi, WANG Yue BAI, Miao ZHANG, Ming-lu , REN Lian-hai. Isolation, Identification and Degradation Characteristics of Functional Bacteria for NH3 and H2S Degradation [J]. Biotechnology Bulletin, 2023, 39(9): 246-254. |
| [4] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [5] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [6] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [7] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [8] | MA Jun-xiu, WU Hao-qiong, JIANG Wei, YAN Geng-xuan, HU Ji-hua, ZHANG Shu-mei. Screening and Identification of Broad-spectrum Antagonistic Bacterial Strains Against Vegetable Soft Rot Pathogen and Its Control Effects [J]. Biotechnology Bulletin, 2023, 39(7): 228-240. |
| [9] | XIE Dong, WANG Liu-wei, LI Ning-jian, LI Ze-lin, XU Zi-hang, ZHANG Qing-hua. Exploration, Identification and Phosphorus-solubilizing Condition Optimization of a Multifunctional Strain [J]. Biotechnology Bulletin, 2023, 39(7): 241-253. |
| [10] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [11] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [12] | LI Xin-yi, JIANG Chun-xiu, XUE Li, JIANG Hong-tao, YAO Wei, DENG Zu-hu, ZHANG Mu-qing, YU Fan. Enhancing Hybridization Signal of Sugarcane Chromosome Oligonucleotide Probe via Multiple Fluorescence Labeled Primers [J]. Biotechnology Bulletin, 2023, 39(5): 103-111. |
| [13] | WANG Yi-fan, HOU Lin-hui, CHANG Yong-chun, YANG Ya-jie, CHEN Tian, ZHAO Zhu-yue, RONG Er-hua, WU Yu-xiang. Synthesis and Character Identification of Allohexaploid Between Gossypium hirsutum and G. gossypioides [J]. Biotechnology Bulletin, 2023, 39(5): 168-176. |
| [14] | CHE Yong-mei, GUO Yan-ping, LIU Guang-chao, YE Qing, LI Ya-hua, ZHAO Fang-gui, LIU Xin. Isolation and Identification of Bacterial Strain C8 and B4 and Their Halotolerant Growth-promoting Effects and Mechanisms [J]. Biotechnology Bulletin, 2023, 39(5): 276-285. |
| [15] | CHEN Xiao-meng, ZHANG Xue-jing, ZHANG Huan, ZHANG Bao-jiang, SU Yan. Prokaryotic Expression of Recombinant Bovine Mastitis Staphylococcus aureus GapC Protein and Identification of Its B-cell Epitopes [J]. Biotechnology Bulletin, 2023, 39(5): 306-313. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||