








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (2): 269-280.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0453
Previous Articles Next Articles
LUO Xue-cong( ), AN Meng-nan, WU Yuan-hua, XIA Zi-hao(
), AN Meng-nan, WU Yuan-hua, XIA Zi-hao( )
)
Received:2021-04-07
Online:2022-02-26
Published:2022-03-09
Contact:
XIA Zi-hao
E-mail:1193645288@qq.com;zihao8337@syau.edu.cn
LUO Xue-cong, AN Meng-nan, WU Yuan-hua, XIA Zi-hao. Applications of Recombinase Polymerase Amplification in Plant Virus Detection[J]. Biotechnology Bulletin, 2022, 38(2): 269-280.
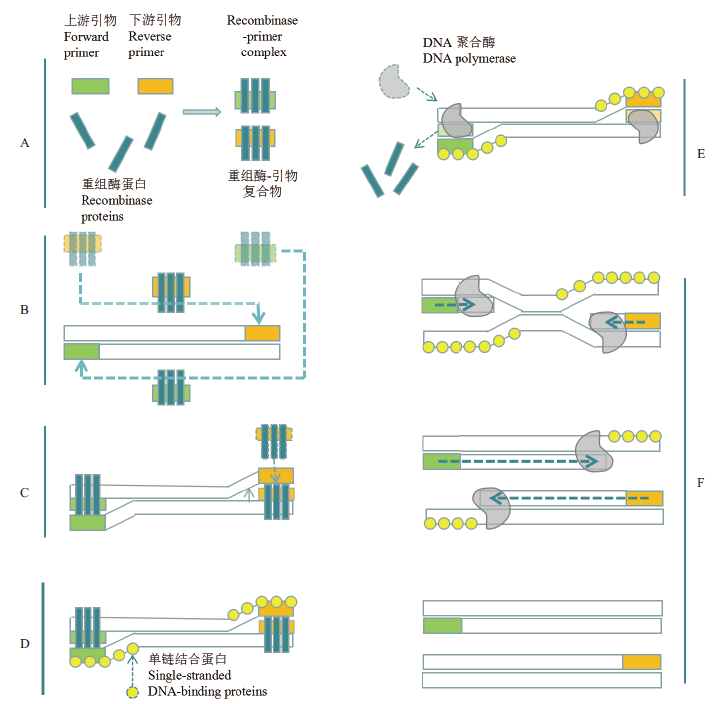
Fig. 1 Schematic diagram of RPA principle[11] A:Formation of recombinant enzyme-primer complex. B:Homologous sequences on localization templates. C:Formation of D-loop structure by initiation of chain exchange reaction. D:Single-stranded DNA-binding proteins stabilize the replaced DNA chain. E:DNA polymerases identify the exposed 3'-end of the primers after dissociation of the recombinant enzyme. F:DNA amplification
| 病毒 Virus | 英文全称 Full name in English | 缩写 Acronym | 属 Genus | 方法 Method | 模板 Template | 灵敏度 Sensitivity | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| 苹果茎沟病毒 | Apple stem grooving virus | ASGV | Capillovirus | RT-RPA | 纯化的RNA | 4.7 ng | [ |
| 苹果茎痘病毒 | Apple stem pitting virus | ASPV | Foveavirus | RT-RPA | CP体外转录物 | 10-4稀释度 | [ |
| 迈索尔香蕉线条病毒 | Banana streak Mysore virus | BSMYV | Badnavirus | RT-RPA | 纯化的RNA | 10-2稀释度 | [ |
| 叶片粗提物 | 10-2稀释度 | ||||||
| 大麦黄矮病毒 | Barley yellow dwarf virus | BYDV | Luteovirus | RT-RPA | CP体外转录物 | 10-7稀释度 | [ |
| 菜豆荚斑驳病毒 | Bean pod mottle virus | BPMV | Comovirus | RT-RPA | 总RNA | 10-4 稀释度 | [ |
| IAC-RT-RPA | 总RNA | 0.05 ng | [ | ||||
| 樱桃病毒A | Cherry virus A | CVA | Capillovirus | RT-RPA-LFS | 总RNA | 10-4稀释度 | [ |
| 辣椒脉斑驳病毒 | Chilli veinal mottle virus | ChiVMV | Potyvirus | RT-RPA | 总RNA | 10 fg | [ |
| 柑橘衰退病毒 | Citrus tristeza virus | CTV | Closterovirus | RT-RPA-AuNP-EIS | CTV-p20 gene | 1 pg | [ |
| RT-RPA-LFICA | 总RNA | 3.77×105拷贝 | [ | ||||
| RNA体外转录物 | 6.288×108拷贝 | ||||||
| 黄瓜花叶病毒 | Cucumber mosaic virus | CMV | Cucumovirus | RT-exoRPA | 纯化的RNA | 3 pg | [ |
| 叶片粗提物 | 10-4稀释度 | ||||||
| 纯化的RNA | 3 pg | ||||||
| 叶片粗提物 | 10-5稀释度 | ||||||
| 黄瓜绿斑驳花叶病毒 | Cucumber green mottle mosaic virus | CGMMV | Tobamovirus | RT-RPA | 总RNA | 10-6稀释度 | [ |
| RT-RPA | 总RNA | 0.5 pg | [ | ||||
| 南瓜黄矮化失调病毒 | Cucurbit yellow stunting disorder virus | CYSDV | Crinivirus | RT-exoRPA | 纯化的总RNA | 10拷贝 | [ |
| 葡萄卷叶伴随病毒2号 | Grapevine leafroll-associated virus 2 | GLRaV-2 | Ampelovirus | RT-RPA | 总RNA | 10-4稀释度 | [ |
| 葡萄卷叶伴随病毒3号 | Grapevine leafroll-associated virus 3 | GLRaV-3 | Ampelovirus | RT-RPA | 总RNA | 10-3稀释度 | [ |
| 莴苣花叶病毒 | Lettuce mosaic virus | LMV | Potyvirus | RT-RPA | 总RNA | 10-2 稀释度 | [ |
| 小樱桃病毒2号 | Little cherry virus 2 | LChV2 | Ampelovirus | RT-RPA-LFS | 叶片粗提物 | 10-2稀释度 | [ |
| 纯化的RNA | 0.1 ng | ||||||
| 莲藕潜隐病毒 | Lotus latent virus | LLV | Potyvirus | RT-RPA | 质粒 | 7x103拷贝 | [ |
| 玉米褪绿斑驳病毒 | Maize chlorotic mottle virus | MCMV | Machlomovirus | RT-RPA | 总RNA | 2.3×10-6 μg | [ |
| 总RNA | 10-4稀释度 | [ | |||||
| 胡椒黄斑驳病毒 | Pepper yellow mottle virus | PYMoV | Badnavirus | RPA | 总DNA | 10-5稀释度 | [ |
| 粗提物 | 10-3稀释度 | ||||||
| RT-RPA | 总RNA | 10-5稀释度 | |||||
| 李痘病毒 | Plum pox virus | PPV | Potyvirus | RT-RPA-LFS | 叶片粗提物 | 10-4稀释度 | [ |
| RNA体外转录物 | 1 fg | ||||||
| real-time RT-RPA | 叶片粗提物 | 10-4稀释度 | |||||
| RNA体外转录物 | 16 fg | ||||||
| RT-RPA | 总RNA | 1.06 ng | [ | ||||
| 马铃薯Y病毒 | Potato virus Y | PVY | Potyvirus | RT-RPA | 纯化的总RNA | 10-2稀释度 | [ |
| 叶片粗提物 | 10-2稀释度 | ||||||
| real-time RT-RPA | 纯化的总RNA | 10-3稀释度 | |||||
| 李矮缩病毒 | Prune dwarf virus | PDV | Ilarvirus | RPA-LFD | 总RNA | 170 fg | [ |
| 水稻黑条矮缩病毒 | Rice black-streaked dwarf virus | RBSDV | Fijivirus | RT-RPA-LFS | 总RNA | 10-1稀释度 | [ |
| 玫瑰丛簇病毒 | Rose rosette virus | RRV | Emaravirus | RT-exoRPA | RNA体外转录物 | 1 fg | [ |
| RT-RPA | RNA体外转录物 | 1 fg | [ | ||||
| 南方菜豆花叶病毒 | Southern bean mosaic virus | SBMV | Sobemovirus | RT-RPA | 总RNA | 10-3稀释度 | [ |
| IAC-RT-RPA | 总RNA | 0.05 ng | [ | ||||
| 大豆花叶病毒 | Soybean mosaic virus | SMV | Potyvirus | RT-RPA | 总RNA | 10-4稀释度 | [ |
| IAC-RT-RPA | 总RNA | 0.05 ng | [ | ||||
| 番茄褪绿病毒 | Tomato chlorosis virus | ToCV | Crinivirus | RT-RPA | 总RNA | 3 ng | [ |
| 番茄斑萎病毒 | Tomato spotted wilt virus | TSWV | Tospovirus | RT-RPA | 总RNA | 10-5稀释度 | [ |
| 淮山药温和花叶病毒 | Yam mild mosaic virus | YMMV | Potyvirus | Real-time RT-RPA | 叶片粗提物 | 10-1稀释度 | [ |
| 薯蓣花叶病毒 | Yam mosaic virus | YMV | Potyvirus | RT-exoRPA | 纯化的RNA | 14 pg | [ |
| Real-time RT-RPA | 叶片粗提物 | 10-3稀释度 | [ | ||||
| 香蕉束顶病毒 | Banana bunchy top virus | BBTV | Babuvirus | RPA | 基因组DNA | 10-6稀释度 | [ |
| 叶片粗提物 | 10-6稀释度 | ||||||
| 菜豆金色黄花叶病毒 | Bean golden yellow mosaic virus | BGYMV | Begomovirus | RPA | 叶片粗提物 | 10-3稀释度 | [ |
| 柑橘黄花叶病毒 | Citrus yellow mosaic virus | CYMV | Badnavirus | RPA | 叶片粗提物 | 10-5稀释度 | [ |
| IC-RPA | 叶片粗提物 | - | |||||
| 紫云英矮缩病毒 | Milk vetch dwarf virus | MDV | Nanovirus | RPA-LFS | 质粒 | 10拷贝 | [ |
| RPA | 质粒 | 10拷贝 | |||||
| 番茄斑驳病毒 | Tomato mottle virus | ToMoV | Begomovirus | RPA | 叶片粗提物 | 10-3稀释度 | [ |
| 番茄黄化曲叶病毒 | Tomato yellow leaf curl virus | TYLCV | Begomovirus | RPA | 总DNA | 50 pg | [ |
| RPA-AuNP | 总DNA | 1拷贝 | [ | ||||
| RPA | 质粒 | 9.6 pg | [ | ||||
| 粗提物 | 10-3稀释度 | ||||||
| 啤酒花矮化类病毒 | Hop stunt viroid | HSVd | Hostuviroid | RT-RPA-LFS | 总RNA | 10 ng | [ |
| RNA体外转录物 | 2×109拷贝 | ||||||
| RT-RPA | 基因组RNA | - | [ | ||||
| 桃潜隐花叶类病毒 | Peach latent mosaic viroid | PLMVd | Pelamoviroid | RT-RPA | 总RNA | 10-7稀释度 | [ |
| 马铃薯纺锤块茎类病毒 | Potato spindle tuber viroid | PSTVd | Pospiviroid | RT-RPA-LFA | RNA体外转录物 | 106拷贝 | [ |
| 总RNA | 10-7稀释度 | ||||||
| 番茄褪绿矮缩类病毒 | Tomato chlorotic dwarf viroid | TCDVd | Pospiviroid | RT-RPA-LFS | RNA体外转录物 | 1 pg | [ |
| 叶片粗提物 | 25倍稀释 | ||||||
| 种子粗提物 | 10-1稀释度 |
Table 1 Application of RPA in plant virus detection
| 病毒 Virus | 英文全称 Full name in English | 缩写 Acronym | 属 Genus | 方法 Method | 模板 Template | 灵敏度 Sensitivity | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| 苹果茎沟病毒 | Apple stem grooving virus | ASGV | Capillovirus | RT-RPA | 纯化的RNA | 4.7 ng | [ |
| 苹果茎痘病毒 | Apple stem pitting virus | ASPV | Foveavirus | RT-RPA | CP体外转录物 | 10-4稀释度 | [ |
| 迈索尔香蕉线条病毒 | Banana streak Mysore virus | BSMYV | Badnavirus | RT-RPA | 纯化的RNA | 10-2稀释度 | [ |
| 叶片粗提物 | 10-2稀释度 | ||||||
| 大麦黄矮病毒 | Barley yellow dwarf virus | BYDV | Luteovirus | RT-RPA | CP体外转录物 | 10-7稀释度 | [ |
| 菜豆荚斑驳病毒 | Bean pod mottle virus | BPMV | Comovirus | RT-RPA | 总RNA | 10-4 稀释度 | [ |
| IAC-RT-RPA | 总RNA | 0.05 ng | [ | ||||
| 樱桃病毒A | Cherry virus A | CVA | Capillovirus | RT-RPA-LFS | 总RNA | 10-4稀释度 | [ |
| 辣椒脉斑驳病毒 | Chilli veinal mottle virus | ChiVMV | Potyvirus | RT-RPA | 总RNA | 10 fg | [ |
| 柑橘衰退病毒 | Citrus tristeza virus | CTV | Closterovirus | RT-RPA-AuNP-EIS | CTV-p20 gene | 1 pg | [ |
| RT-RPA-LFICA | 总RNA | 3.77×105拷贝 | [ | ||||
| RNA体外转录物 | 6.288×108拷贝 | ||||||
| 黄瓜花叶病毒 | Cucumber mosaic virus | CMV | Cucumovirus | RT-exoRPA | 纯化的RNA | 3 pg | [ |
| 叶片粗提物 | 10-4稀释度 | ||||||
| 纯化的RNA | 3 pg | ||||||
| 叶片粗提物 | 10-5稀释度 | ||||||
| 黄瓜绿斑驳花叶病毒 | Cucumber green mottle mosaic virus | CGMMV | Tobamovirus | RT-RPA | 总RNA | 10-6稀释度 | [ |
| RT-RPA | 总RNA | 0.5 pg | [ | ||||
| 南瓜黄矮化失调病毒 | Cucurbit yellow stunting disorder virus | CYSDV | Crinivirus | RT-exoRPA | 纯化的总RNA | 10拷贝 | [ |
| 葡萄卷叶伴随病毒2号 | Grapevine leafroll-associated virus 2 | GLRaV-2 | Ampelovirus | RT-RPA | 总RNA | 10-4稀释度 | [ |
| 葡萄卷叶伴随病毒3号 | Grapevine leafroll-associated virus 3 | GLRaV-3 | Ampelovirus | RT-RPA | 总RNA | 10-3稀释度 | [ |
| 莴苣花叶病毒 | Lettuce mosaic virus | LMV | Potyvirus | RT-RPA | 总RNA | 10-2 稀释度 | [ |
| 小樱桃病毒2号 | Little cherry virus 2 | LChV2 | Ampelovirus | RT-RPA-LFS | 叶片粗提物 | 10-2稀释度 | [ |
| 纯化的RNA | 0.1 ng | ||||||
| 莲藕潜隐病毒 | Lotus latent virus | LLV | Potyvirus | RT-RPA | 质粒 | 7x103拷贝 | [ |
| 玉米褪绿斑驳病毒 | Maize chlorotic mottle virus | MCMV | Machlomovirus | RT-RPA | 总RNA | 2.3×10-6 μg | [ |
| 总RNA | 10-4稀释度 | [ | |||||
| 胡椒黄斑驳病毒 | Pepper yellow mottle virus | PYMoV | Badnavirus | RPA | 总DNA | 10-5稀释度 | [ |
| 粗提物 | 10-3稀释度 | ||||||
| RT-RPA | 总RNA | 10-5稀释度 | |||||
| 李痘病毒 | Plum pox virus | PPV | Potyvirus | RT-RPA-LFS | 叶片粗提物 | 10-4稀释度 | [ |
| RNA体外转录物 | 1 fg | ||||||
| real-time RT-RPA | 叶片粗提物 | 10-4稀释度 | |||||
| RNA体外转录物 | 16 fg | ||||||
| RT-RPA | 总RNA | 1.06 ng | [ | ||||
| 马铃薯Y病毒 | Potato virus Y | PVY | Potyvirus | RT-RPA | 纯化的总RNA | 10-2稀释度 | [ |
| 叶片粗提物 | 10-2稀释度 | ||||||
| real-time RT-RPA | 纯化的总RNA | 10-3稀释度 | |||||
| 李矮缩病毒 | Prune dwarf virus | PDV | Ilarvirus | RPA-LFD | 总RNA | 170 fg | [ |
| 水稻黑条矮缩病毒 | Rice black-streaked dwarf virus | RBSDV | Fijivirus | RT-RPA-LFS | 总RNA | 10-1稀释度 | [ |
| 玫瑰丛簇病毒 | Rose rosette virus | RRV | Emaravirus | RT-exoRPA | RNA体外转录物 | 1 fg | [ |
| RT-RPA | RNA体外转录物 | 1 fg | [ | ||||
| 南方菜豆花叶病毒 | Southern bean mosaic virus | SBMV | Sobemovirus | RT-RPA | 总RNA | 10-3稀释度 | [ |
| IAC-RT-RPA | 总RNA | 0.05 ng | [ | ||||
| 大豆花叶病毒 | Soybean mosaic virus | SMV | Potyvirus | RT-RPA | 总RNA | 10-4稀释度 | [ |
| IAC-RT-RPA | 总RNA | 0.05 ng | [ | ||||
| 番茄褪绿病毒 | Tomato chlorosis virus | ToCV | Crinivirus | RT-RPA | 总RNA | 3 ng | [ |
| 番茄斑萎病毒 | Tomato spotted wilt virus | TSWV | Tospovirus | RT-RPA | 总RNA | 10-5稀释度 | [ |
| 淮山药温和花叶病毒 | Yam mild mosaic virus | YMMV | Potyvirus | Real-time RT-RPA | 叶片粗提物 | 10-1稀释度 | [ |
| 薯蓣花叶病毒 | Yam mosaic virus | YMV | Potyvirus | RT-exoRPA | 纯化的RNA | 14 pg | [ |
| Real-time RT-RPA | 叶片粗提物 | 10-3稀释度 | [ | ||||
| 香蕉束顶病毒 | Banana bunchy top virus | BBTV | Babuvirus | RPA | 基因组DNA | 10-6稀释度 | [ |
| 叶片粗提物 | 10-6稀释度 | ||||||
| 菜豆金色黄花叶病毒 | Bean golden yellow mosaic virus | BGYMV | Begomovirus | RPA | 叶片粗提物 | 10-3稀释度 | [ |
| 柑橘黄花叶病毒 | Citrus yellow mosaic virus | CYMV | Badnavirus | RPA | 叶片粗提物 | 10-5稀释度 | [ |
| IC-RPA | 叶片粗提物 | - | |||||
| 紫云英矮缩病毒 | Milk vetch dwarf virus | MDV | Nanovirus | RPA-LFS | 质粒 | 10拷贝 | [ |
| RPA | 质粒 | 10拷贝 | |||||
| 番茄斑驳病毒 | Tomato mottle virus | ToMoV | Begomovirus | RPA | 叶片粗提物 | 10-3稀释度 | [ |
| 番茄黄化曲叶病毒 | Tomato yellow leaf curl virus | TYLCV | Begomovirus | RPA | 总DNA | 50 pg | [ |
| RPA-AuNP | 总DNA | 1拷贝 | [ | ||||
| RPA | 质粒 | 9.6 pg | [ | ||||
| 粗提物 | 10-3稀释度 | ||||||
| 啤酒花矮化类病毒 | Hop stunt viroid | HSVd | Hostuviroid | RT-RPA-LFS | 总RNA | 10 ng | [ |
| RNA体外转录物 | 2×109拷贝 | ||||||
| RT-RPA | 基因组RNA | - | [ | ||||
| 桃潜隐花叶类病毒 | Peach latent mosaic viroid | PLMVd | Pelamoviroid | RT-RPA | 总RNA | 10-7稀释度 | [ |
| 马铃薯纺锤块茎类病毒 | Potato spindle tuber viroid | PSTVd | Pospiviroid | RT-RPA-LFA | RNA体外转录物 | 106拷贝 | [ |
| 总RNA | 10-7稀释度 | ||||||
| 番茄褪绿矮缩类病毒 | Tomato chlorotic dwarf viroid | TCDVd | Pospiviroid | RT-RPA-LFS | RNA体外转录物 | 1 pg | [ |
| 叶片粗提物 | 25倍稀释 | ||||||
| 种子粗提物 | 10-1稀释度 |
| [1] | 李大伟. 我国植物病毒学的研究现状及发展策略[J]. 植物保护, 2010, 36(2):5-8. |
| Li DW. Current status and development strategy for plant virology in China[J]. Plant Prot, 2010, 36(2):5-8. | |
| [2] | 商明清, 魏梅生. 植物病毒检测新技术研究进展[J]. 植物检疫, 2004, 18(4):236-240. |
| Shang MQ, Wei MS. Advances in new technologies for plant virus detection[J]. Plant Quar, 2004, 18(4):236-240. | |
| [3] | 谢晓亮. 丹参病毒病原鉴定与脱病毒技术研究[D]. 北京:北京林业大学, 2008. |
| Xie XL. Studies on identification of viral disease pathogens and establishment of virus-free technology in Salvia miltiorrhiza bge[D]. Beijing:Beijing Forestry University, 2008. | |
| [4] | 施曼玲, 周雪平. 植物病毒病的诊断技术[J]. 微生物学通报, 2000, 27(2):149-151. |
| Shi ML, Zhou XP. Diagnostic techniques for plant viral diseases[J]. Microbiology, 2000, 27(2):149-151. | |
| [5] | 韩彤. 沈阳三种木本植物病毒病的分子检测与鉴定[D]. 沈阳:沈阳大学, 2018. |
| Han T. Detection and identification of three woody plant virus diseases in Shenyang[D]. Shenyang:Shenyang University, 2018. | |
| [6] |
Cheung VG, Morley M, Aguilar F, et al. Making and reading microarrays[J]. Nat Genet, 1999, 21(1 Suppl):15-19.
pmid: 9915495 |
| [7] |
Babu B, Ochoa-Corona FM, Paret ML. Recombinase polymerase amplification applied to plant virus detection and potential implications[J]. Anal Biochem, 2018, 546:72-77.
doi: 10.1016/j.ab.2018.01.021 URL |
| [8] |
Piepenburg O, Williams CH, Stemple DL, et al. DNA detection using recombination proteins[J]. PLoS Biol, 2006, 4(7):e204.
doi: 10.1371/journal.pbio.0040204 URL |
| [9] |
Boyle DS, McNerney R, Teng Low H, et al. Rapid detection of Mycobacterium tuberculosis by recombinase polymerase amplification[J]. PLoS One, 2014, 9(8):e103091.
doi: 10.1371/journal.pone.0103091 URL |
| [10] |
景志刚, 董浩, 狄栋栋, 等. 重组酶聚合酶扩增技术研究进展[J]. 生物技术通报, 2016, 32(6):47-53.
doi: 10.13560/j.cnki.biotech.bull.1985.2016.06.008 |
| Jing ZG, Dong H, Di DD, et al. Research progress on recombinase polymerase amplification[J]. Biotechnol Bull, 2016, 32(6):47-53. | |
| [11] | 施奕, 徐昌平, 余蓓蓓, 等. 重组酶聚合酶扩增技术研究进展[J]. 病毒学报, 2020, 36(3):522-532. |
| Shi Y, Xu CP, Yu BB, et al. Research progress in recombinase polymerase amplification(RPA)[J]. Chin J Virol, 2020, 36(3):522-532. | |
| [12] |
Sharma N, Hoshika S, Hutter D, et al. Recombinase-based isothermal amplification of nucleic acids with self-avoiding molecular recognition systems(SAMRS)[J]. Chembiochem, 2014, 15(15):2268-2274.
doi: 10.1002/cbic.201402250 pmid: 25209570 |
| [13] |
Wee EJH, Trau M. Simple isothermal strategy for multiplexed, rapid, sensitive, and accurate miRNA detection[J]. ACS Sens, 2016, 1(6):670-675.
doi: 10.1021/acssensors.6b00105 URL |
| [14] | 杜亚楠, 赵笑, 范小瑞, 等. 重组酶聚合酶扩增技术的研究进展及其应用[J]. 上海农业学报, 2018, 34(6):117-122. |
| Du YN, Zhao X, Fan XR, et al. Advances and applications of recombinase polymerase amplification[J]. Acta Agric Shanghai, 2018, 34(6):117-122. | |
| [15] | 张娜, 乾义柯, 魏霜, 等. 基于重组酶聚合酶扩增技术(RPA)的葡萄卷叶伴随病毒3号检测方法[J]. 新疆农业科学, 2016, 53(2):302-308. |
| Zhang N, Qian YK, Wei S, et al. Based on Recombinase Polymerase Amplification, the Method of Detection of Grapevine leafroll-associated virus 3[J]. Xinjiang Agric Sci, 2016, 53(2):302-308. | |
| [16] |
Henson JM, French R. The polymerase chain reaction and plant disease diagnosis[J]. Annu Rev Phytopathol, 1993, 31:81-109.
pmid: 18643762 |
| [17] |
Jiao Y, Jiang J, An M, et al. Recombinase polymerase amplification assay for rapid detection of maize chlorotic mottle virus in maize[J]. Arch Virol, 2019, 164(10):2581-2584.
doi: 10.1007/s00705-019-04361-3 URL |
| [18] |
Nassuth A, Pollari E, Helmeczy K, et al. Improved RNA extraction and one-tube RT-PCR assay for simultaneous detection of control plant RNA plus several viruses in plant extracts[J]. J Virol Methods, 2000, 90(1):37-49.
pmid: 11011079 |
| [19] | 周莹, 岳瑾, 李云龙, 等. 重组酶介导的等温扩增技术及其在植物病原检测中的应用[J]. 中国植保导刊, 2020, 40(6):27-31. |
| Zhou Y, Yue J, Li YL, et al. Recombinase polymerase amplification and its applications in plant pathogens detection[J]. China Plant Prot, 2020, 40(6):27-31. | |
| [20] | 陈玲, 闫国华, 张晓明, 等. 李矮缩病毒重组酶聚合酶扩增—侧流层析试纸条检测方法的建立[J]. 园艺学报, 2021, 48(1):183-192. |
| Chen L, Yan GH, Zhang XM, et al. Establishment of recombinase polymerase amplification combined with lateral flow dipstick for detection of prune dwarf virus[J]. Acta Hortic Sin, 2021, 48(1):183-192. | |
| [21] | 薛佳莹, 崔向红, 曹涤非, 等. 测流层析技术标记材料的研究进展[J]. 化学工程师, 2020, 34(11):52-54, 57. |
| Xue JY, Cui XH, Cao DF, et al. Progress in label materials of lateral flow chromatography[J]. Chem Eng, 2020, 34(11):52-54, 57. | |
| [22] | 马兰, 王淑娟, 曾海娟, 等. 侧流层析技术研究进展[J]. 食品科学, 2018, 39(15):333-342. |
| Ma L, Wang SJ, Zeng HJ, et al. Progress in lateral flow chromatography[J]. Food Sci, 2018, 39(15):333-342. | |
| [23] |
Li J, Macdonald J, von Stetten F. Review:a comprehensive summary of a decade development of the recombinase polymerase amplification[J]. Analyst, 2018, 144(1):31-67.
doi: 10.1039/C8AN01621F URL |
| [24] |
Cao Y, Yan D, Wu X, et al. Rapid and visual detection of milk vetch dwarf virus using recombinase polymerase amplification combined with lateral flow strips[J]. Virol J, 2020, 17(1):102.
doi: 10.1186/s12985-020-01371-5 URL |
| [25] | 仇有文, 张明辉, 高学军, 等. Taqman定量PCR技术检测转基因大豆中外源基因拷贝数[J]. 安徽农业科学, 2011, 39(17):10150-10152. |
| Qiu YW, Zhang MH, Gao XJ, et al. Detection of foreign gene copies in transgenic soybean by taqman quantitative PCR technique[J]. J Anhui Agric Sci, 2011, 39(17):10150-10152. | |
| [26] |
Euler M, Wang YJ, Heidenreich D, et al. Development of a panel of recombinase polymerase amplification assays for detection of biothreat agents[J]. J Clin Microbiol, 2013, 51(4):1110-1117.
doi: 10.1128/JCM.02704-12 URL |
| [27] |
Srivastava N, Kapoor R, Kumar R, et al. Rapid diagnosis of Cucumber mosaic virus in banana plants using a fluorescence-based real-time isothermal reverse transcription-recombinase polymerase amplification assay[J]. J Virol Methods, 2019, 270:52-58.
doi: S0166-0934(18)30330-6 pmid: 31047971 |
| [28] |
Babu B, Washburn BK, Ertek TS, et al. A field based detection method for Rose rosette virus using isothermal probe-based Reverse transcription-recombinase polymerase amplification assay[J]. J Virol Methods, 2017, 247:81-90.
doi: 10.1016/j.jviromet.2017.05.019 URL |
| [29] |
Ahmed A, van der Linden H, Hartskeerl RA. Development of a recombinase polymerase amplification assay for the detection of pathogenic Leptospira[J]. Int J Environ Res Public Health, 2014, 11(5):4953-4964.
doi: 10.3390/ijerph110504953 URL |
| [30] |
Fang WF, Chen WJ, Yang JT. Colorimetric determination of DNA concentration and mismatches using hybridization-mediated growth of gold nanoparticle probes[J]. Sens Actuat B:Chem, 2014, 192:77-82.
doi: 10.1016/j.snb.2013.10.052 URL |
| [31] |
Wang TM, Yang JT. Visual DNA diagnosis of Tomato yellow leaf curl virus with integrated recombinase polymerase amplification and a gold-nanoparticle probe[J]. Sci Rep, 2019, 9(1):15146.
doi: 10.1038/s41598-019-51650-7 URL |
| [32] |
Wang Z, Zhang J, Ekman JM, et al. DNA-mediated control of metal nanoparticle shape:one-pot synjournal and cellular uptake of highly stable and functional gold nanoflowers[J]. Nano Lett, 2010, 10(5):1886-1891.
doi: 10.1021/nl100675p URL |
| [33] |
Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system[J]. Nat Protoc, 2013, 8(11):2281-2308.
doi: 10.1038/nprot.2013.143 URL |
| [34] |
Ali Z, Abulfaraj A, Idris A, et al. CRISPR/Cas9-mediated viral interference in plants[J]. Genome Biol, 2015, 16:238.
doi: 10.1186/s13059-015-0799-6 URL |
| [35] |
Mahas A, Aman R, Mahfouz M. CRISPR-Cas13d mediates robust RNA virus interference in plants[J]. Genome Biol, 2019, 20(1):263.
doi: 10.1186/s13059-019-1881-2 URL |
| [36] |
Kellner MJ, Koob JG, Gootenberg JS, et al. SHERLOCK:nucleic acid detection with CRISPR nucleases[J]. Nat Protoc, 2019, 14(10):2986-3012.
doi: 10.1038/s41596-019-0210-2 pmid: 31548639 |
| [37] |
Aman R, Mahas A, Marsic T, et al. Efficient, rapid, and sensitive detection of plant RNA viruses with one-pot RT-RPA-CRISPR/Cas12a assay[J]. Front Microbiol, 2020, 11:610872.
doi: 10.3389/fmicb.2020.610872 URL |
| [38] | 张皖静. 直扩RPA核酸即时可视化检测技术研究[D]. 上海:上海师范大学, 2020. |
| Zhang WJ. Study on RPA nucleic acid visual point-of-care test technology[D]. Shanghai:Shanghai Normal University, 2020. | |
| [39] |
Kim NY, Oh J, Lee SH, et al. Rapid and specific detection of Apple stem grooving virus by reverse transcription-recombinase polymerase amplification[J]. Plant Pathol J, 2018, 34(6):575-579.
doi: 10.5423/PPJ.NT.06.2018.0108 URL |
| [40] |
Kim NY, Lee HJ, Jeong RD. A portable detection assay for Apple stem pitting virus using reverse transcription-recombinase polymerase amplification[J]. J Virol Methods, 2019, 274:113747.
doi: 10.1016/j.jviromet.2019.113747 URL |
| [41] |
Kapoor R, Srivastava N, Kumar R, et al. Detection of episomal banana streak Mysore virus by reverse transcription-recombinase polymerase amplification assay[J]. J Plant Pathol, 2020, 102(2):499-503.
doi: 10.1007/s42161-019-00424-1 URL |
| [42] |
Kim NK, Kim SM, Jeong RD. Reverse transcription recombinase polymerase amplification assay for rapid and sensitive detection of barley yellow dwarf virus in oat[J]. Plant Pathol J, 2020, 36(5):497-502.
doi: 10.5423/PPJ.NT.08.2020.0148 URL |
| [43] | 张永江, 魏霜, 袁俊杰, 等. 一步法逆转录重组酶聚合酶常温扩增(RT-RPA)技术检测菜豆荚斑驳病毒[J]. 江苏农业科学, 2018, 46(21):96-98. |
| Zhang YJ, Wei S, Yuan JJ, et al. One-step reverse transcription recombinase polymerase amplification(RT-RPA)technology for detection of BPMV[J]. Jiangsu Agric Sci, 2018, 46(21):96-98. | |
| [44] | 袁俊杰, 龙阳, 渭婷玉, 等. 添加扩增内标的逆转录重组酶聚合酶扩增技术检测三种大豆病毒[J]. 农业生物技术学报, 2020, 28(12):2261-2269. |
| Yuan JJ, Long Y, Wei TY, et al. RT-RPA technology included with IAC for detection of three soybean(Glycine max)viruses[J]. J Agric Biotechnol, 2020, 28(12):2261-2269. | |
| [45] | 陈玲, 段续伟, 张开春, 等. 基于重组酶聚合酶扩增(RPA)技术的樱桃病毒A(CVA)的检测方法[J]. 园艺学报, 2020, 47(2):390-398. |
| Chen L, Duan XW, Zhang KC, et al. A method for the detection of cherry virus A(CVA)based on recombinase polymerase amplification(RPA)technique[J]. Acta Hortic Sin, 2020, 47(2):390-398. | |
| [46] | Jiao YB, Xu CT, Li JL, et al. Characterization and a RT-RPA assay for rapid detection of Chilli veinal mottle virus(ChiVMV)in tobacco[J]. virol J, 2020, 17(1):33. |
| [47] |
Khater M, Escosura-Muñiz A, Altet L, et al. In situ plant virus nucleic acid isothermal amplification detection on gold nanoparticle-modified electrodes[J]. Anal Chem, 2019, 91(7):4790-4796.
doi: 10.1021/acs.analchem.9b00340 pmid: 30843387 |
| [48] |
Ghosh DK, Kokane SB, Gowda S. Development of a reverse transcription recombinase polymerase based isothermal amplification coupled with lateral flow immunochromatographic assay(CTV-RT-RPA-LFICA)for rapid detection of Citrus tristeza virus[J]. Sci Rep, 2020, 10(1):20593.
doi: 10.1038/s41598-020-77692-w URL |
| [49] |
Jiao Y, Jiang J, Wu Y, et al. Rapid detection of Cucumber green mottle mosaic virus in watermelon through a recombinase polymerase amplification assay[J]. J Virol Methods, 2019, 270:146-149.
doi: 10.1016/j.jviromet.2019.05.008 URL |
| [50] |
Zeng R, Luo J, Gao S, et al. Rapid detection of Cucumber green mottle mosaic virus by reverse transcription recombinase polymerase amplification[J]. Mol Cell Probes, 2019, 43:84-85.
doi: 10.1016/j.mcp.2018.12.005 URL |
| [51] |
Kalischuk ML, Roberts PD, Paret ML. A rapid fluorescence-based real-time isothermal assay for the detection of Cucurbit yellow stunting disorder virus in squash and watermelon plants[J]. Mol Cell Probes, 2020, 53:101613.
doi: 10.1016/j.mcp.2020.101613 URL |
| [52] | 张永江, 魏霜, 黄帅, 等. 中国植物病理学会2017年学术年会论文集[C]. 山东: 中国农业科学技术出版社, 2017: 7. |
| Zhang YJ, Wei S, Huang S, et al. Proceedings of the 2017 Annual Conference of the Chinese Plant Pathology Society[C]. Shandong: China Agricultural Science and Technology Press, 2017: 7. | |
| [53] | 向均, 王垚, 文朝慧, 等. 莴苣花叶病毒RT-RPA检测方法的建立[J]. 植物检疫, 2018, 32(4):48-51. |
| Xiang J, Wang Y, Wen ZH, et al. Reverse transcription recombinase polymerase amplification assay for the detection of Lettuce mosic virus[J]. Plant Quar, 2018, 32(4):48-51. | |
| [54] |
Mekuria TA, Zhang S, Eastwell KC. Rapid and sensitive detection of Little cherry virus 2 using isothermal reverse transcription-recombinase polymerase amplification[J]. J Virol Methods, 2014, 205:24-30.
doi: 10.1016/j.jviromet.2014.04.015 pmid: 24797461 |
| [55] | 吴伟文. 实时荧光定量和等温核酸扩增检测莲藕潜隐病毒技术的建立与应用[D]. 扬州:扬州大学, 2019. |
| Wu WW. Establishment and application of real-time quantitative PCR and isothermal nucleic acid amplification for detection of Lotus latent virus[D]. Yangzhou:Yangzhou University, 2019. | |
| [56] |
Jiao Y, Jiang J, An M, et al. Recombinase polymerase amplification assay for rapid detection of maize chlorotic mottle virus in maize[J]. Arch Virol, 2019, 164(10):2581-2584.
doi: 10.1007/s00705-019-04361-3 URL |
| [57] | 冯黎霞, 魏霜, 余辛, 等. 重组酶聚合酶扩增技术(RPA)快速检测玉米褪绿斑驳病毒[J]. 植物保护学报, 2020, 47(1):217-218. |
| Feng LX, Wei S, Yu X, et al. Detection of maize chlorotic mottle virus by recombinase polymerase amplification[J]. J Plant Prot, 2020, 47(1):217-218. | |
| [58] |
Mohandas A, Bhat AI. Recombinase polymerase amplification assay for the detection of Piper yellow mottle virus infecting black pepper[J]. Virusdisease, 2020, 31(1):38-44.
doi: 10.1007/s13337-019-00566-x pmid: 32206697 |
| [59] |
Zhang SL, Ravelonandro M, Russell P, et al. Rapid diagnostic detection of plum pox virus in Prunus plants by isothermal AmplifyRP® using reverse transcription-recombinase polymerase amplification[J]. J Virol Methods, 2014, 207:114-120.
doi: 10.1016/j.jviromet.2014.06.026 URL |
| [60] | 霍亚云. 四种重要植物病原物恒温扩增检测技术的建立[D]. 北京:中国农业科学院, 2018. |
| Huo YY. Development of detection methods for four important plant pathogens on the base of isothermal amplification[D]. Beijing:Chinese Academy of Agricultural Sciences, 2018. | |
| [61] |
Babujee L, Witherell RA, Mikami K, et al. Optimization of an isothermal recombinase polymerase amplification method for real-time detection of Potato virus Y O and N types in potato[J]. J Virol Methods, 2019, 267:16-21.
doi: 10.1016/j.jviromet.2019.02.006 URL |
| [62] |
Zhao C, Sun F, Li X, et al. Reverse transcription-recombinase polymerase amplification combined with lateral flow strip for detection of rice black-streaked dwarf virus in plants[J]. J Virol Methods, 2019, 263:96-100.
doi: 10.1016/j.jviromet.2018.11.001 URL |
| [63] |
Babu B, Washburn BK, Miller SH, et al. A rapid assay for detection of Rose rosette virus using reverse transcription-recombinase polymerase amplification using multiple gene targets[J]. J Virol Methods, 2017, 240:78-84.
doi: 10.1016/j.jviromet.2016.11.014 URL |
| [64] | 魏霜, 袁俊杰, 李桂芬, 等. 南方菜豆花叶病毒RT-RPA检测方法的建立[J]. 植物检疫, 2018, 32(1):50-53. |
| Wei S, Yuan JJ, Li GF, et al. One-step reverse transcription recombinase polymerase amplification(RT-RPA)for the detection of Southern bean mosaic virus[J]. Plant Quar, 2018, 32(1):50-53. | |
| [65] | 袁俊杰, 魏霜, 龙阳, 等. 一步法逆转录重组酶聚合酶常温扩增技术(RT-RPA)检测大豆花叶病毒[J]. 检验检疫学刊, 2018, 28(2):1-4. |
| Yuan JJ, Wei S, Long Y, et al. One-step reverse transcription recombinase polymerase amplification(RT-RPA)for the detection of SMV[J]. J Insp Quar, 2018, 28(2):1-4. | |
| [66] | 宋建, 薛俊, 孙海波, 等. 一种基于RPA的番茄褪绿病毒检测方法[J]. 植物保护, 2020, 46(4):168-170, 184. |
| Song J, Xue J, Sun HB, et al. Detection of Tomato chlorotic virus based on RPA[J]. Plant Prot, 2020, 46(4):168-170, 184. | |
| [67] | 余辛, 魏梅生, 林晓红, 等. 番茄斑萎病毒RT-RPA检测方法的建立[J]. 植物检疫, 2020, 34(5):46-49. |
| Yu X, Wei MS, Lin XH, et al. Development of RT-RPA for detection of tomato spotted wilt virus[J]. Plant Quar, 2020, 34(5):46-49. | |
| [68] |
Silva G, Oyekanmi J, Nkere CK, et al. Rapid detection of potyviruses from crude plant extracts[J]. Anal Biochem, 2018, 546:17-22.
doi: 10.1016/j.ab.2018.01.019 URL |
| [69] |
Silva G, Bömer M, Nkere C, et al. Rapid and specific detection of Yam mosaic virus by reverse-transcription recombinase polymerase amplification[J]. J Virol Methods, 2015, 222:138-144.
doi: 10.1016/j.jviromet.2015.06.011 URL |
| [70] |
Kapoor R, Srivastava N, Kumar S, et al. Development of a recombinase polymerase amplification assay for the diagnosis of banana bunchy top virus in different banana cultivars[J]. Arch Virol, 2017, 162(9):2791-2796.
doi: 10.1007/s00705-017-3399-9 URL |
| [71] |
Londoño MA, Harmon CL, Polston JE. Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics[J]. Virol J, 2016, 13:48.
doi: 10.1186/s12985-016-0504-8 pmid: 27000806 |
| [72] |
Kumar PV, Sharma SK, Rishi N, et al. An isothermal based recombinase polymerase amplification assay for rapid, sensitive and robust indexing of Citrus yellow mosaic virus[J]. Acta virol, 2018, 62(1):104-108.
doi: 10.4149/av_2018_113 pmid: 29521109 |
| [73] | 周莹, 杨丽梅, 刘梅, 等. 重组酶聚合酶扩增技术在番茄黄化曲叶病毒检测中的应用[J]. 中国蔬菜, 2019(1):36-40. |
| Zhou Y, Yang LM, Liu M, et al. Application of recombinase polymerase amplification technology in detecting TYLCV[J]. China Veg, 2019(1):36-40. | |
| [74] |
Kappagantu M, Villamor DEV, Bullock JM, et al. A rapid isothermal assay for the detection of Hop stunt viroid in hop plants(Humulus lupulus), and its application in disease surveys[J]. J Virol Methods, 2017, 245:81-85.
doi: S0166-0934(16)30417-7 pmid: 28392409 |
| [75] | Stackhouse T, Waliullah S, Oliver JE, et al. First report of hop stunt viroid infecting Citrus trees in Georgia, USA[J]. Plant Dis, 2021, 105(2):515. |
| [76] |
Lee HJ, Kim HJ, Lee K, et al. Rapid detection of peach latent mosaic viroid by reverse transcription recombinase polymerase amplification[J]. Mol Cell Probes, 2020, 53:101627.
doi: 10.1016/j.mcp.2020.101627 URL |
| [77] |
Ivanov AV, Shmyglya IV, Zherdev AV, et al. The challenge for rapid detection of high-structured circular RNA:assay of potato spindle Tuber viroid based on recombinase polymerase amplification and lateral flow tests[J]. Plants, 2020, 9(10):1369.
doi: 10.3390/plants9101369 URL |
| [78] |
Hammond RW, Zhang SL. Development of a rapid diagnostic assay for the detection of tomato chlorotic dwarf viroid based on isothermal reverse-transcription-recombinase polymerase amplification[J]. J Virol Methods, 2016, 236:62-67.
doi: 10.1016/j.jviromet.2016.06.013 URL |
| [1] | LI Huan-min, GAO Feng-tao, LI Wei-zhong, WANG Jin-qing, FENG Jia-li. Progress in Research and Application of Natural Bio-materials as Immobilized Carriers [J]. Biotechnology Bulletin, 2023, 39(7): 105-112. |
| [2] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [3] | ZHANG Xue-ping, LU Yu-qing, ZHANG Yue-qian, LI Xiao-juan. Advances in Plant Extracellular Vesicles and Analysis Techniques [J]. Biotechnology Bulletin, 2023, 39(5): 32-43. |
| [4] | WANG Xiao-mei, YANG Xiao-wei, LI Hui-shang, HE Wei, XIN Zhu-lin. Development Status of Synthetic Biology in Globe and Its Enlightenment [J]. Biotechnology Bulletin, 2023, 39(2): 292-302. |
| [5] | ZHOU Xi-wen, CHENG Ke, ZHU Hong-liang. Research Progress in the Approaches to in vivo RNA Secondary Structure Profiling in Plants [J]. Biotechnology Bulletin, 2023, 39(2): 51-62. |
| [6] | GUO Wen-bo, LU Yang, SUI Li, ZHAO Yu, ZOU Xiao-wei, ZHANG Zheng-kun, LI Qi-yun. Preparation and Application of Polyclonal Antibodies Against Beauveria bassiana Mycovirus BbPmV-4 Coat Protein [J]. Biotechnology Bulletin, 2023, 39(10): 58-67. |
| [7] | LI Hui-jie, DONG Lian-hua, CHEN Gui-fang, LIU Si-yuan, YANG Jia-yi, YANG Jing-ya. Establishment of Droplet Digital PCR Assay for Quantitative Detection of Pseudomonas cocovenenans in Foods [J]. Biotechnology Bulletin, 2023, 39(1): 127-136. |
| [8] | CHEN Xiao-lin, LIU Yang-er, XU Wen-tao, GUO Ming-zhang, LIU Hui-lin. Application of Synthetic Biology Based Whole-cell Biosensor Technology in the Rapid Detection of Food Safety [J]. Biotechnology Bulletin, 2023, 39(1): 137-149. |
| [9] | HU Hai-yang, YING Wan-qin, HE Jun, LV Zhi-xian, XIE Xiao-ping, DENG Zhong-liang. Establishment and Application of ERA Real-time Fluorescence Method for Rapid Detection of Mycoplasma pneumoniae [J]. Biotechnology Bulletin, 2022, 38(9): 264-270. |
| [10] | GAO Wei-xin, HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng. Construction and Activity Verification of Ribonucleoprotein Complex for Gene Editing [J]. Biotechnology Bulletin, 2022, 38(8): 60-68. |
| [11] | SUN De-quan, LU Xin-hua, LI Wei-ming, HU Yu-lin, DUAN Ya-jie, PANG Zhen-cai, HU Hui-gang. Application of Mesoporous Silica Nanoparticles in Agriculture [J]. Biotechnology Bulletin, 2022, 38(5): 228-239. |
| [12] | XIONG He-li, SHA Qian, LIU Shao-na, XIANG De-cai, ZHANG Bin, ZHAO Zhi-yong. Application of Single-cell Transcriptome Sequencing in Animals [J]. Biotechnology Bulletin, 2022, 38(3): 226-233. |
| [13] | LAN Xin-yue, LIU Ning-ning, ZHU Long-jiao, CHEN Xu, CHU Hua-shuo, LI Xiang-yang, DUAN Nuo, XU Wen-tao. Tetracycline Bivalent Aptamer Non-enzyme Label-free Sensor [J]. Biotechnology Bulletin, 2022, 38(3): 276-284. |
| [14] | KONG De-zhen, NIE Ying-bin, XU Hong-jun, CUI Feng-juan, MU Pei-yuan, TIAN Xiao-ming. Effects of Blend Seeding on the Yield,Purity and Yield Advantage of F1 in Three-line Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(10): 132-139. |
| [15] | CHEN Li, LU Xi, YANG Hong-lan, ZHANG Peng, HE Zhi-xu. Methodological Research on Rapid Detection of CRISPR/Cas9-Mediated Gene Mutations in Cells Based on High-resolution Melting Technique [J]. Biotechnology Bulletin, 2022, 38(10): 90-96. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||