








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (3): 1-8.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0499
ZHOU Juan( ), YAN Jin-dong, LI Xin-mei, LIU Xue-qing, ZHAO Qiang, ZHAO Xiao-ying(
), YAN Jin-dong, LI Xin-mei, LIU Xue-qing, ZHAO Qiang, ZHAO Xiao-ying( )
)
Received:2021-04-16
Online:2022-03-26
Published:2022-04-06
Contact:
ZHAO Xiao-ying
E-mail:2395498678@qq.com;xiaoyzhao@hnu.edu.cn
ZHOU Juan, YAN Jin-dong, LI Xin-mei, LIU Xue-qing, ZHAO Qiang, ZHAO Xiao-ying. Study on the Interaction of F-box Protein FKF1 and Transcription Factor FUL in Regulating Flowering in Arabidopsis[J]. Biotechnology Bulletin, 2022, 38(3): 1-8.
| 基因座Locus | 蛋白名称Name of protein | 分值Score | 功能Function |
|---|---|---|---|
| AT1G22770 | Gigantea protein(GI) | 0.954 | Long day pathway |
| AT1G04400 | Cryptochrome 2(CRY2) | 0.952 | Light perception |
| AT5G60910 | AGAMOUS-like 8(FUL) | 0.946 | Floral promoter |
| AT3G42830 | RING/U-box superfamily protein(RBX1b) | 0.938 | |
| AT2G45660 | AGAMOUS-like 20(SOC1) | 0.9 | Floral promoter |
| AT5G20570 | RING-box 1(RBX1a) | 0.876 | |
| AT5G15840 | B-box type zinc finger protein with CCT domain(CO/BBX1) | 0.87 | Long day pathway |
| AT1G10940 | Protein kinase superfamily protein(SNRK1A) | 0.766 | |
| AT2G32950 | Transducin/WD40 repeat-like superfamily protein(COP1) | 0.726 | |
| AT1G09570 | Phytochrome A(PhyA) | 0.614 | Light perception |
| AT2G18790 | Phytochrome B(PhyB) | 0.57 | Light perception |
| AT5G11260 | Basic-leucine zipper(bZIP)transcription factor family protein(HY5) | 0.512 | |
| AT4G16250 | Phytochrome D(PhyD) | 0.87 | Light perception |
| AT2G25930 | Hydroxyproline-rich glycoprotein family protein(ELF3) | 0.878 | Circadian clock |
| AT2G46830 | Circadian clock associated 1(CCA1) | 0.608 | Circadian clock |
| AT1G09530 | Phytochrome interacting factor 3(PIF3) | 0.6 | Light signaling |
| AT5G61380 | CCT motif-containing response regulator protein(TOC1) | 0.888 | Circadian clock |
Table 1 Candidate interaction proteins of FKF1
| 基因座Locus | 蛋白名称Name of protein | 分值Score | 功能Function |
|---|---|---|---|
| AT1G22770 | Gigantea protein(GI) | 0.954 | Long day pathway |
| AT1G04400 | Cryptochrome 2(CRY2) | 0.952 | Light perception |
| AT5G60910 | AGAMOUS-like 8(FUL) | 0.946 | Floral promoter |
| AT3G42830 | RING/U-box superfamily protein(RBX1b) | 0.938 | |
| AT2G45660 | AGAMOUS-like 20(SOC1) | 0.9 | Floral promoter |
| AT5G20570 | RING-box 1(RBX1a) | 0.876 | |
| AT5G15840 | B-box type zinc finger protein with CCT domain(CO/BBX1) | 0.87 | Long day pathway |
| AT1G10940 | Protein kinase superfamily protein(SNRK1A) | 0.766 | |
| AT2G32950 | Transducin/WD40 repeat-like superfamily protein(COP1) | 0.726 | |
| AT1G09570 | Phytochrome A(PhyA) | 0.614 | Light perception |
| AT2G18790 | Phytochrome B(PhyB) | 0.57 | Light perception |
| AT5G11260 | Basic-leucine zipper(bZIP)transcription factor family protein(HY5) | 0.512 | |
| AT4G16250 | Phytochrome D(PhyD) | 0.87 | Light perception |
| AT2G25930 | Hydroxyproline-rich glycoprotein family protein(ELF3) | 0.878 | Circadian clock |
| AT2G46830 | Circadian clock associated 1(CCA1) | 0.608 | Circadian clock |
| AT1G09530 | Phytochrome interacting factor 3(PIF3) | 0.6 | Light signaling |
| AT5G61380 | CCT motif-containing response regulator protein(TOC1) | 0.888 | Circadian clock |
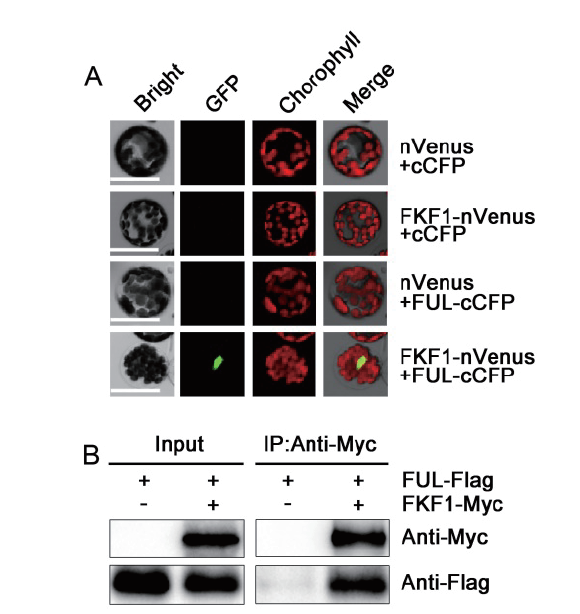
Fig. 2 BiFC and Co-IP assays showing FKF1 interaction with FUL A:BiFC assay showing the interaction of FKF1 with FUL in nucleus in Arabidopsis protoplast. Bar = 50μm. B:Co-IP assay showing the interaction of FKF1 with FUL
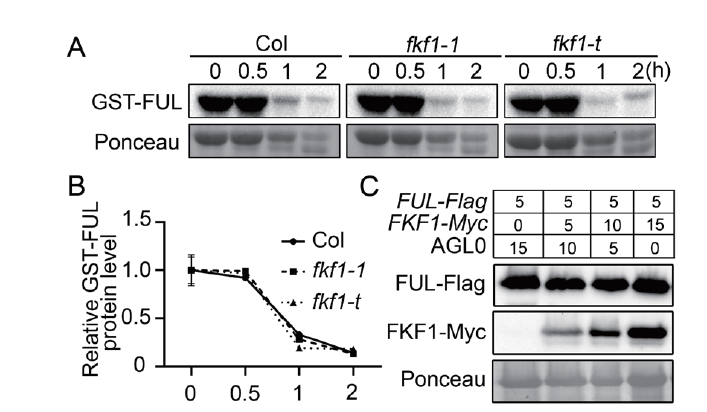
Fig. 3 Effect of FKF1 on FUL protein stability A:Semi-in vivo stability analysis of FUL protein. Escherichia coli-purified GST-FUL protein was mixed with total protein from the 12 d seedlings of wild-type Col,fkf1-1 or fkf1-t in equal proportion,and incubated for the given time. Sampled,and anti-GST antibody was detected by Western blotting. Ponceau staining was used as an internal standard. Three independent experiments were conducted,showing similar results. B:Protein level of GST-FUL in(A). GST-FUL protein level was normalized to Ponceau. The value of the starting point was set to 1. Bars refer to the standard deviations of three biological replicates. C:In vivo stability analysis of FUL protein. The FUL-Flag was co-expressed with increasing amounts of FKF1-Myc in tobacco leaves,and the total protein was extracted. The anti-Flag and anti-Myc proteins were detected by Western blotting,respectively. Numbers indicate the ratios of the concentrations of Agrobacteria used in co-infiltration. Ponceau staining was used as an internal standard. Three independent experiments were conducted,showing similar results
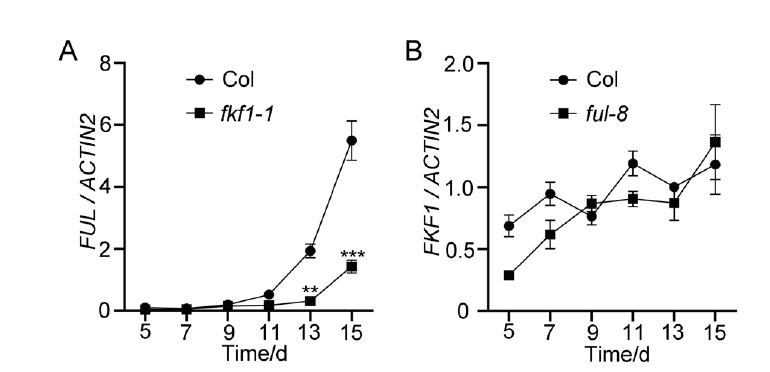
Fig. 4 FUL mRNA expression regulated by FKF1 A:mRNA expression of FUL in the wild-type Col and fkf1-1 mutant. B:mRNA expression of FKF1 in the wild-type Col and ful-8 mutant. Plants grew on 1/2 MS solid medium under long-day conditions(LDs),and aboveground seedlings were collected for quantitative real-time PCR(qRT-PCR)analysis. ACTIN2 served as the internal control. Error bars refer to the standard deviations of three biological replicates. Significant differences are indicated:**P<0.01,***P<0.001(Tukey's least significant difference test)
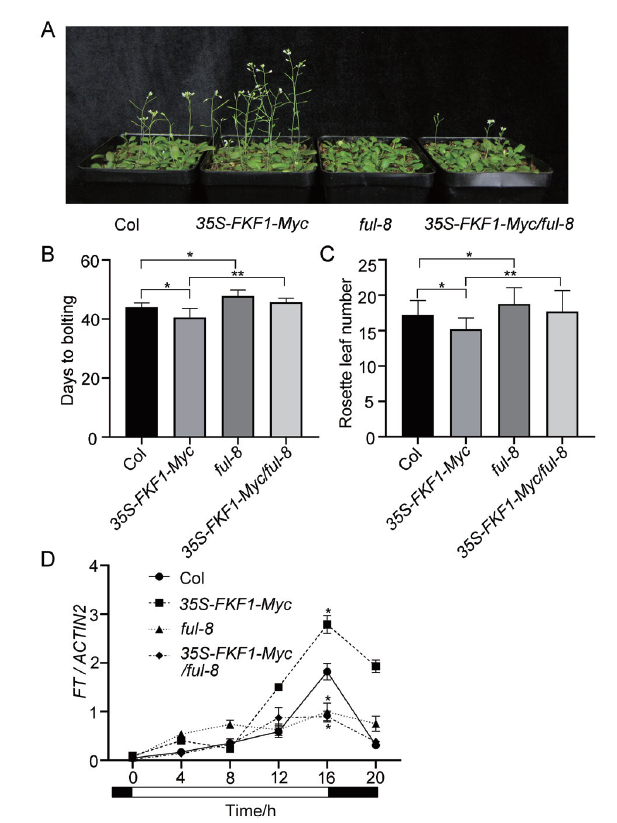
Fig. 5 Flowering phenotype of Col,35S-FKF1-Myc,ful-8 and 35S-FKF1-Myc / ful-8 plants A:Images of 40 d plants grown in soil under LDs. B:The days of plant sowed to bolting and blossoming of the respective genotypes. C:Number of rosette leaves at bolting of the respective genotypes. D:mRNA expression levels of FT in the respective genotypes. Plants grew in 1/2MS solid medium under LDs,samples were collected at every 4 h on day 17 for qRT-PCR analysis. ACTIN2 served as the internal control. Error bars refers to the standard deviations of three biological replicates. The white/black bars refer to light/dark phases. The time of collecting first sample is set as 0 h. Error bar refers to standard deviations(n≥10). Significant differences are indicated:*P<0.05 and **P<0.01(Tukey's least significant difference test)
| [1] |
Bernier G, Périlleux C. A physiological overview of the genetics of flowering time control[J]. Plant Biotechnol J, 2005, 3(1):3-16.
doi: 10.1111/pbi.2005.3.issue-1 URL |
| [2] | Fornara F, de Montaigu A, Coupland G. SnapShot:control of flowering in Arabidopsis[J]. Cell, 2010,141(3):550-550. e2. |
| [3] |
Srikanth A, Schmid M. Regulation of flowering time:all roads lead to Rome[J]. Cell Mol Life Sci, 2011, 68(12):2013-2037.
doi: 10.1007/s00018-011-0673-y pmid: 21611891 |
| [4] |
Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development[J]. Trends Genet, 2010, 26(12):519-527.
doi: 10.1016/j.tig.2010.09.001 pmid: 20947199 |
| [5] |
Kardailsky I, Shukla VK, Ahn JH, et al. Activation tagging of the floral inducer FT[J]. Science, 1999, 286(5446):1962-1965.
pmid: 10583961 |
| [6] |
Lee J, Oh M, Park H, et al. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy[J]. Plant J, 2008, 55(5):832-843.
doi: 10.1111/tpj.2008.55.issue-5 URL |
| [7] |
Zhang L, Chen L, Yu D. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering[J]. Plant Physiol, 2018, 176(1):790-803.
doi: 10.1104/pp.17.00657 URL |
| [8] |
Lee H, Suh SS, Park E, et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis[J]. Genes Dev, 2000, 14(18):2366-2376.
doi: 10.1101/gad.813600 URL |
| [9] |
Yamaguchi A, Kobayashi Y, Goto K, et al. TWIN SISTER OF FT(TSF)Acts as a floral pathway integrator redundantly with FT[J]. Plant Cell Physiol, 2005, 46(8):1175-1189.
pmid: 15951566 |
| [10] |
Jang S, Torti S, Coupland G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis[J]. Plant J, 2009, 60(4):614-625.
doi: 10.1111/tpj.2009.60.issue-4 URL |
| [11] |
Suárez-López P, Wheatley K, Robson F, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis[J]. Nature, 2001, 410(6832):1116-1120.
doi: 10.1038/35074138 URL |
| [12] |
Valverde F. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering[J]. Science, 2004, 303(5660):1003-1006.
doi: 10.1126/science.1091761 URL |
| [13] |
Wigge PA. Integration of spatial and temporal information during floral induction in Arabidopsis[J]. Science, 2005, 309(5737):1056-1059.
doi: 10.1126/science.1114358 URL |
| [14] |
Imaizumi T, Tran HG, Swartz TE, et al. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis[J]. Nature, 2003, 426(6964):302-306.
doi: 10.1038/nature02090 URL |
| [15] |
Nelson DC, Lasswell J, Rogg LE, et al. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis[J]. Cell, 2000, 101(3):331-340.
pmid: 10847687 |
| [16] |
Imaizumi T, Schultz TF, Harmon FG, et al. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis[J]. Science, 2005, 309(5732):293-297.
pmid: 16002617 |
| [17] |
Sawa M, Nusinow DA, Kay SA, et al. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis[J]. Science, 2007, 318(5848):261-265.
doi: 10.1126/science.1146994 URL |
| [18] |
Song YH, Smith RW, To BJ, et al. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering[J]. Science, 2012, 336(6084):1045-1049.
doi: 10.1126/science.1219644 URL |
| [19] |
Lee BD, Kim MR, Kang MY, et al. The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering[J]. Nat Commun, 2017, 8(1):2259.
doi: 10.1038/s41467-017-02476-2 URL |
| [20] |
Lee BD, Cha JY, Kim MR, et al. Light-dependent suppression of COP1 multimeric complex formation is determined by the blue-light receptor FKF1 in Arabidopsis[J]. Biochem Biophys Res Commun, 2019, 508(1):191-197.
doi: 10.1016/j.bbrc.2018.11.032 URL |
| [21] |
Yan J, Li X, Zeng B, et al. FKF1 F-box protein promotes flowering in part by negatively regulating DELLA protein stability under long-day photoperiod in Arabidopsis[J]. J Integr Plant Biol, 2020, 62(11):1717-1740.
doi: 10.1111/jipb.v62.11 URL |
| [22] |
Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana[J]. Cell, 2009, 138(4):738-749.
doi: 10.1016/j.cell.2009.06.014 URL |
| [23] |
Ferrándiz C, Gu Q, Martienssen R, et al. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER[J]. Development, 2000, 127(4):725-734.
pmid: 10648231 |
| [24] |
Melzer S, Lens F, Gennen J, et al. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana[J]. Nat Genet, 2008, 40(12):1489-1492.
doi: 10.1038/ng.253 URL |
| [25] |
Mandel MA, Yanofsky MF. The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1[J]. Plant Cell, 1995, 7(11):1763-1771.
pmid: 8535133 |
| [26] |
Gu Q, Ferrándiz C, Yanofsky MF, et al. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development[J]. Development, 1998, 125(8):1509-1517.
pmid: 9502732 |
| [27] |
Schmid M, Uhlenhaut NH, Godard F, et al. Dissection of floral induction pathways using global expression analysis[J]. Development, 2003, 130(24):6001-6012.
doi: 10.1242/dev.00842 URL |
| [28] |
Abe M, Kobayashi Y, Yamamoto S, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex[J]. Science, 2005, 309(5737):1052-1056.
doi: 10.1126/science.1115983 URL |
| [29] |
Torti S, Fornara F, Vincent C, et al. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering[J]. Plant Cell, 2012, 24(2):444-462.
doi: 10.1105/tpc.111.092791 URL |
| [30] |
Yamaguchi A, Wu MF, Yang L, et al. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1[J]. Dev Cell, 2009, 17(2):268-278.
doi: 10.1016/j.devcel.2009.06.007 pmid: 19686687 |
| [31] |
Li W, Wang HP, Yu DQ. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions[J]. Mol Plant, 2016, 9(11):1492-1503.
doi: 10.1016/j.molp.2016.08.003 URL |
| [32] |
Hyun Y, Richter R, Vincent C, et al. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem[J]. Dev Cell, 2016, 37(3):254-266.
doi: 10.1016/j.devcel.2016.04.001 URL |
| [33] |
Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts:a versatile cell system for transient gene expression analysis[J]. Nat Protoc, 2007, 2(7):1565-1572.
doi: 10.1038/nprot.2007.199 URL |
| [34] |
Sparkes IA, Runions J, Kearns A, et al. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants[J]. Nat Protoc, 2006, 1(4):2019-2025.
pmid: 17487191 |
| [35] |
Moon J, Lee H, Kim M, et al. Analysis of flowering pathway integrators in Arabidopsis[J]. Plant Cell Physiol, 2005, 46(2):292-299.
doi: 10.1093/pcp/pci024 URL |
| [36] |
Oda A, Fujiwara S, Kamada H, et al. Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression[J]. FEBS Lett, 2004, 557(1/2/3):259-264.
doi: 10.1016/S0014-5793(03)01470-4 URL |
| [37] |
Kim JJ, Lee JH, Kim W, et al. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis[J]. Plant Physiol, 2012, 159(1):461-478.
doi: 10.1104/pp.111.192369 URL |
| [38] |
Lee JH, Yoo SJ, Park SH, et al. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis[J]. Genes Dev, 2007, 21(4):397-402.
doi: 10.1101/gad.1518407 URL |
| [39] |
Mathieu J, Yant LJ, Mürdter F, et al. Repression of flowering by the miR172 target SMZ[J]. PLoS Biol, 2009, 7(7):e1000148.
doi: 10.1371/journal.pbio.1000148 URL |
| [40] |
Searle I, He Y, Turck F, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis[J]. Genes Dev, 2006, 20(7):898-912.
doi: 10.1101/gad.373506 URL |
| [41] |
Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering[J]. Curr Biol, 2008, 18(17):1338-1343.
doi: 10.1016/j.cub.2008.07.075 pmid: 18718758 |
| [42] |
Liu H, Yu X, Li K, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis[J]. Science, 2008, 322(5907):1535-1539.
doi: 10.1126/science.1163927 URL |
| [43] |
Kumar SV, Lucyshyn D, Jaeger KE, et al. Transcription factor PIF4 controls the thermosensory activation of flowering[J]. Nature, 2012, 484(7393):242-245.
doi: 10.1038/nature10928 URL |
| [44] |
Aerts N, de Bruijn S, van Mourik H, et al. Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana[J]. BMC Plant Biol, 2018, 18(1):131.
doi: 10.1186/s12870-018-1348-8 URL |
| [45] |
Ito S, Song YH, Imaizumi T. LOV domain-containing F-box proteins:light-dependent protein degradation modules in Arabidopsis[J]. Mol Plant, 2012, 5(3):573-582.
doi: 10.1093/mp/sss013 URL |
| [1] | WANG Bao-bao, WANG Hai-yang. Molecular Design of Ideal Plant Architecture for High-density Tolerance of Maize Plant [J]. Biotechnology Bulletin, 2023, 39(8): 11-30. |
| [2] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [3] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [4] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [5] | CUI Ji-jie, CAI Wen-bo, ZHUANG Qing-hui, GAO Ai-ping, HUANG Jian-feng, CHEN Ya-hui, SONG Zhi-zhong. Biological Function of Gene MiISU1 for Fe-S Cluster Assembly in Mangifera indica [J]. Biotechnology Bulletin, 2023, 39(2): 139-146. |
| [6] | HOU Rui-ze BAO Yue CHEN Qi-liang MAO Gui-ling WEI Bo-lin HOU Lei-ping LI Mei-lan. Cloning,Expression and Functional Identification of PRR5 Gene in Pakchoi [J]. Biotechnology Bulletin, 2023, 39(10): 128-135. |
| [7] | RUAN Hang, DUO Hao-yuan, FAN Wen-yan, LV Qing-han, JIANG Shu-jun, ZHU Sheng-wei. Role of the AtERF49 in the Responses to Salt-alkali Stress in Arabidopsis [J]. Biotechnology Bulletin, 2023, 39(1): 150-156. |
| [8] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [9] | GAO Cong, XIAO Chu-jian, LU Shuai, WANG Su-rong, YUAN Hui-hua, CAO Yun-ying. Promoting Effect of Graphene Oxide on the Root Growth of Arabidopsis thaliana [J]. Biotechnology Bulletin, 2022, 38(6): 120-128. |
| [10] | XU Hong-yun, ZHANG Ming-yi. AtSCL4,an Arabidopsis thaliana GRAS Transcription Factor,Negatively Modulates Plants in Response to Osmotic Stress [J]. Biotechnology Bulletin, 2022, 38(6): 129-135. |
| [11] | GU Pan, QI Xue-ying, LI Li, ZHANG Xi, SHAN Xiao-yi. Endocytosis of AtRGS1 Involved in the Regulation of G-protein-mediated Arabidopsis Development and Stress Responses [J]. Biotechnology Bulletin, 2022, 38(6): 34-42. |
| [12] | YANG Jia-hui, SUN Yu-ping, LU Ya-ning, LIU huan, LU Cun-fu, CHEN Yu-zhen. Abiotic Stress Resistance of Escherichia coli Transformed with Arabidopsis thaliana AtTERT Gene [J]. Biotechnology Bulletin, 2022, 38(2): 1-9. |
| [13] | LI Bing-juan, ZHENG Lu, SHEN Ren-fang, LAN Ping. Proteomic Analysis of RPP1A Involved in the Seedling Growth of Arabidopsis thaliana [J]. Biotechnology Bulletin, 2022, 38(2): 10-20. |
| [14] | ZHANG Lin, WEI Zhen-zhen, SONG Cheng-wei, GUO Li-li, GUO Qi, HOU Xiao-gai, WANG Hua-fang. Cloning and Expression Analysis of PoFD Gene from Paeonia ostii ‘Fengdan’ [J]. Biotechnology Bulletin, 2022, 38(11): 104-111. |
| [15] | XU Zi-han, LIU Qian, MIAO Da-peng, CHEN Yue, HU Feng-rong. Impacts of Cymbidium goeringii’s miR396 Overexpression on the Leaf Growth,Photosynthesis and Chlorophyll Fluorescence in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2021, 37(5): 28-37. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||