








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (6): 129-135.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0963
Previous Articles Next Articles
Received:2021-07-28
Online:2022-06-26
Published:2022-07-11
Contact:
XU Hong-yun
E-mail:xhyplant@126.com
XU Hong-yun, ZHANG Ming-yi. AtSCL4,an Arabidopsis thaliana GRAS Transcription Factor,Negatively Modulates Plants in Response to Osmotic Stress[J]. Biotechnology Bulletin, 2022, 38(6): 129-135.
| 引物名称 Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| AtSCL4-F | ATGGCTTACATGTGCACTGATAG |
| AtSCL4-R | TTATCGCCAGGAAGAAAGAGTG |
| ATMYB61-F | GATTGCGTCAAGGCTTCCG |
| ATMYB61-R | CGCAGAAGAGGAACTAGGAG |
| P5CS1-F | GTCAAGTCTATGCTTGATTTG |
| P5CS1-R | GATTTGTCGCCGAATGTAATC |
| BADH-F | GCTGACCTAGCTGAAGGCTTG |
| BADH-R | CACCTCGCGGCAAATATCAGC |
| SOD1-F | GTCCACATTTCAACCCCGATG |
| SOD1-R | GAGACCAATGATGCCGCAAGC |
| PER4-F | GAAGGTTGGTCGAAGAGATTC |
| PER4-R | CGTATCTCCACCGTTGACCGG |
| Act7-F | ATGTTCACCACTACCGCAG |
| Act7-R | ACCTCAGGACAACGGAATCTC |
| Tub2-F | GTCTCCAAGGGTTCCAGGTT |
| Tub2-R | GACAGAGTAGCGTTGTAAGGC |
Table 1 Primer sequences used in qRT-PCR
| 引物名称 Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| AtSCL4-F | ATGGCTTACATGTGCACTGATAG |
| AtSCL4-R | TTATCGCCAGGAAGAAAGAGTG |
| ATMYB61-F | GATTGCGTCAAGGCTTCCG |
| ATMYB61-R | CGCAGAAGAGGAACTAGGAG |
| P5CS1-F | GTCAAGTCTATGCTTGATTTG |
| P5CS1-R | GATTTGTCGCCGAATGTAATC |
| BADH-F | GCTGACCTAGCTGAAGGCTTG |
| BADH-R | CACCTCGCGGCAAATATCAGC |
| SOD1-F | GTCCACATTTCAACCCCGATG |
| SOD1-R | GAGACCAATGATGCCGCAAGC |
| PER4-F | GAAGGTTGGTCGAAGAGATTC |
| PER4-R | CGTATCTCCACCGTTGACCGG |
| Act7-F | ATGTTCACCACTACCGCAG |
| Act7-R | ACCTCAGGACAACGGAATCTC |
| Tub2-F | GTCTCCAAGGGTTCCAGGTT |
| Tub2-R | GACAGAGTAGCGTTGTAAGGC |
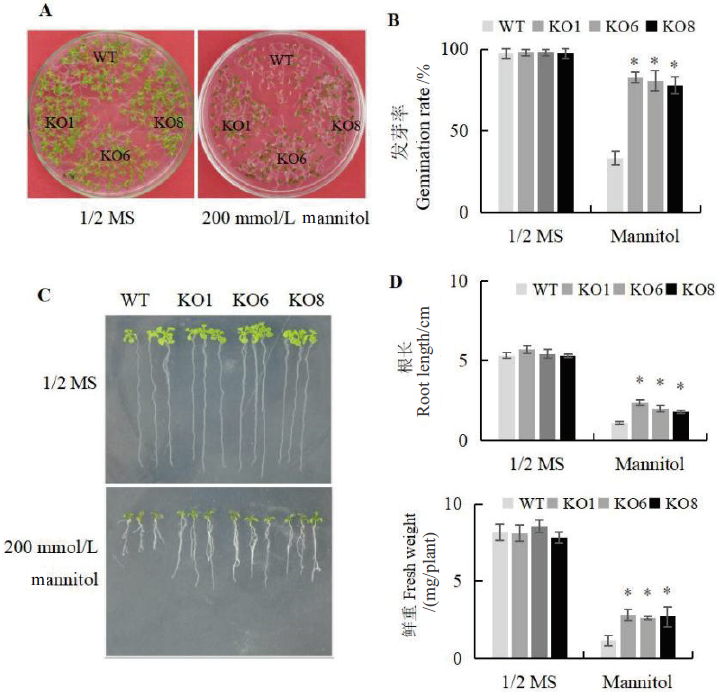
Fig. 3 Analysis of seedling germination and growth regulated by AtSCL4 under osmotic stress A:Observation of the germination phenotypes on plate medium. B:Statistics of germination. C:Observation of root length. D:Statistics of root length and fresh weight
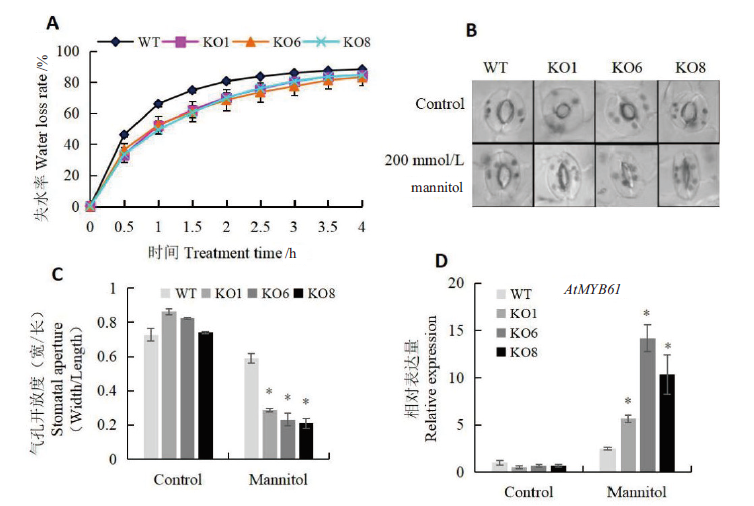
Fig. 4 Stomatal aperture assay of seedlings regulated by AtSCL4 under osmotic stress A:Determination of water loss rate. B:Observation of stomatal aperture.C:Determination of stomatal aperture. D:Expression of AtMYB61 related to stomatal aperture regulation
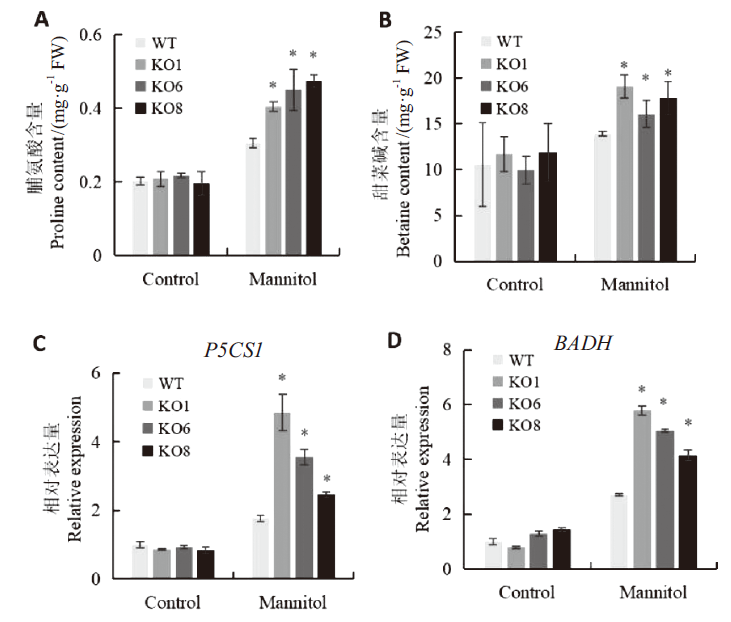
Fig. 5 Synthesis analysis of osmolyte in seedlings mediated by AtSCL4 under osmotic stress A:Proline content. B:Betaine content. C:Expressions of of P5CS1 related to proline synthesis. D:Expressions of BADH related to betaine synthesis
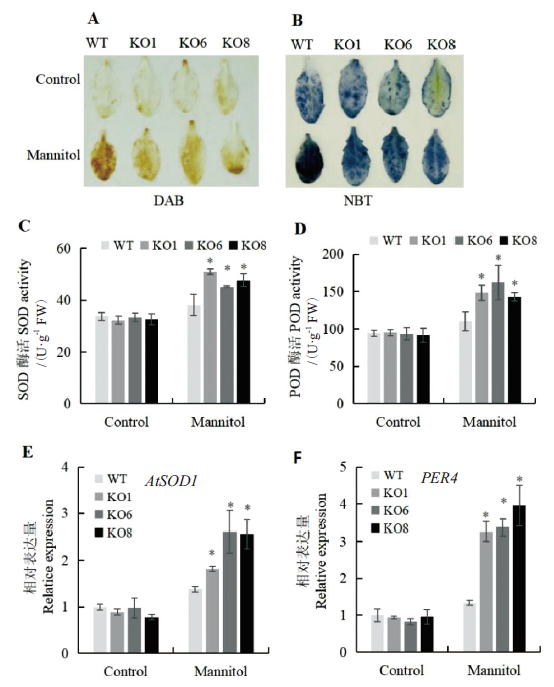
Fig. 6 ROS scavenging capacity assay of seedlings regul-ated by AtSCL4 under osmotic stress A:Observation of H2O2 content by DAB staining. B:Observation of O2-· content by NBT staining. C:SOD activities. D:POD activities. E:Expression of AtSOD1.F:Expression of PER4
| [1] |
Wang J, Song L, Gong X, et al. Functions of jasmonic acid in plant regulation and response to abiotic stress[J]. Int J Mol Sci, 2020, 21(4):1446.
doi: 10.3390/ijms21041446 URL |
| [2] |
Rabbani MA, Maruyama K, Abe H, et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses[J]. Plant Physiol, 2003, 133(4):1755-1767.
pmid: 14645724 |
| [3] |
Hsieh EJ, Cheng MC, Lin TP. Functional characterization of an abiotic stress-inducible transcription factor AtERF53 in Arabidopsis thaliana[J]. Plant Mol Biol, 2013, 82(3):223-237.
doi: 10.1007/s11103-013-0054-z URL |
| [4] |
Sakuraba Y, Kim YS, Han SH, et al. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP[J]. Plant Cell, 2015, 27(6):1771-1787.
doi: 10.1105/tpc.15.00222 URL |
| [5] |
Bolle C. The role of GRAS proteins in plant signal transduction and development[J]. Planta, 2004, 218(5):683-692.
doi: 10.1007/s00425-004-1203-z URL |
| [6] |
Sun X, Xue B, Jones WT, et al. A functionally required unfoldome from the plant kingdom:intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development[J]. Plant Mol Biol, 2011, 77(3):205-223.
doi: 10.1007/s11103-011-9803-z URL |
| [7] |
Liu X, Widmer A. Genome-wide comparative analysis of the GRAS gene family in Populus, Arabidopsis and rice[J]. Plant Mol Biol Report, 2014, 32(6):1129-1145.
doi: 10.1007/s11105-014-0721-5 URL |
| [8] |
Chen YQ, Tai SS, Wang DW, et al. Homology-based analysis of the GRAS gene family in tobacco[J]. Genet Mol Res, 2015, 14(4):15188-15200.
doi: 10.4238/2015.November.25.7 pmid: 26634482 |
| [9] |
Lu J, Wang T, Xu Z, et al. Genome-wide analysis of the GRAS gene family in Prunus mume[J]. Mol Genet Genomics, 2015, 290(1):303-317.
doi: 10.1007/s00438-014-0918-1 URL |
| [10] |
Song XM, Liu TK, Duan WK, et al. Genome-wide analysis of the GRAS gene family in Chinese cabbage(Brassica rapa ssp. pekinensis)[J]. Genomics, 2014, 103(1):135-146.
doi: 10.1016/j.ygeno.2013.12.004 URL |
| [11] |
Huang W, Xian Z, Kang X, et al. Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato[J]. BMC Plant Biol, 2015, 15:209.
doi: 10.1186/s12870-015-0590-6 pmid: 26302743 |
| [12] | Wang Y, Shi S, Zhou Y, et al. Genome-wide identification and characterization of GRAS transcription factors in sacred Lotus(Nelumbo nucifera)[J]. PeerJ, 2016, 4:e2388. |
| [13] |
Xu K, Chen SJ, Li TF, et al. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes[J]. BMC Plant Biol, 2015, 15:141.
doi: 10.1186/s12870-015-0532-3 URL |
| [14] |
Yuan YY, Fang LC, Karungo SK, et al. Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis[J]. Plant Cell Rep, 2016, 35(3):655-666.
doi: 10.1007/s00299-015-1910-x URL |
| [15] |
Ma HS, Liang D, Shuai P, et al. The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana[J]. J Exp Bot, 2010, 61(14):4011-4019.
doi: 10.1093/jxb/erq217 URL |
| [16] |
Zhang S, Li XW, de Fan S, et al. Overexpression of HcSCL13, a Halostachys caspica GRAS transcription factor, enhances plant growth and salt stress tolerance in transgenic Arabidopsis[J]. Plant Physiol Biochem, 2020, 151:243-254.
doi: 10.1016/j.plaphy.2020.03.020 URL |
| [17] | Zhang Y, Zhou YY, Zhang D, et al. PtrWRKY75 overexpression reduces stomatal aperture and improves drought tolerance by salicylic acid-induced reactive oxygen species accumulation in poplar[J]. Environ Exp Bot, 2020, 176:104117. |
| [18] |
Liang YK, Dubos C, Dodd IC, et al. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana[J]. Curr Biol, 2005, 15(13):1201-1206.
doi: 10.1016/j.cub.2005.06.041 URL |
| [19] |
Miller G, Suzuki N, Ciftci-Yilmaz S, et al. Reactive oxygen species homeostasis and signalling during drought and salinity stresses[J]. Plant Cell Environ, 2010, 33(4):453-467.
doi: 10.1111/j.1365-3040.2009.02041.x URL |
| [20] |
Ashraf M, Foolad MR. Roles of Glycine betaine and proline in improving plant abiotic stress resistance[J]. Environ Exp Bot, 2007, 59(2):206-216.
doi: 10.1016/j.envexpbot.2005.12.006 URL |
| [21] |
Silva-Ortega CO, Ochoa-Alfaro AE, Reyes-Agüero JA, et al. Salt stress increases the expression of p5cs gene and induces proline accumulation in Cactus pear[J]. Plant Physiol Biochem, 2008, 46(1):82-92.
doi: 10.1016/j.plaphy.2007.10.011 URL |
| [22] | Ishitani M, Arakawa K, Mizuno K, et al. Betaine aldehyde dehydrogenase in the Gramineae:levels in leaves both betaine-accumulating and nonaccumulating cereal plants[J]. Plant Cell Physiol, 1993, 34(3):493-495. |
| [23] | Petrov V, Hille J, Mueller-Roeber B, et al. ROS-mediated abiotic stress-induced programmed cell death in plants[J]. Front Plant Sci, 2015, 6:69. |
| [24] |
Wang L, Li Z, Lu M, et al. ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis[J]. Front Plant Sci, 2017, 8:635.
doi: 10.3389/fpls.2017.00635 URL |
| [1] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [2] | DING Li, DU Ting-ting, TANG Qiong-ying, GAO Quan-xin, YI Shao-kui, YANG Guo-liang. Analyses of Endocrine Regulation and Expression of Genes Related to the Molting Signaling Pathway in the Molting Cycle of Macrobrachium rosenbergii [J]. Biotechnology Bulletin, 2023, 39(9): 300-310. |
| [3] | LIU Bao-cai, CHEN Jing-ying, ZHANG Wu-jun, HUANG Ying-zhen, ZHAO Yun-qing, LIU Jian-chao, WEI Zhi-cheng. Characteristics Analysis of Seed Microrhizome Gene Expression of Polygonatum cyrtonema [J]. Biotechnology Bulletin, 2023, 39(8): 220-233. |
| [4] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [5] | ZHU Shao-xi, JIN Zhao-yang, GE Jian-rong, WANG Rui, WANG Feng-ge, LU Yun-cai. High-throughput Specific Detection Methods for Transgenic Maize Based on the KASP Platform [J]. Biotechnology Bulletin, 2023, 39(6): 133-140. |
| [6] | MENG Guo-qiang, GUAN Jian-wen, NIU Chun-mei, ZHOU Ying, SHEN Su-lin, WEI You-heng. Construction and Functional Study of RagA Transgenic Drosophila [J]. Biotechnology Bulletin, 2023, 39(6): 171-180. |
| [7] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [8] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [9] | YU Yang, LIU Tian-hai, LIU Li-xu, TANG Jie, PENG Wei-hong, CHEN Yang, TAN Hao. Study on Aerosol Microbial Community in the Production Workshop of Morel Spawn [J]. Biotechnology Bulletin, 2023, 39(5): 267-275. |
| [10] | SONG Hai-na, WU Xin-tong, YANG Lu-yu, GENG Xi-ning, ZHANG Hua-min, SONG Xiao-long. Selection and Validation of Reference Genes for RT-qPCR in Allium tuberosum Infected by Botrytis squamosa [J]. Biotechnology Bulletin, 2023, 39(3): 101-115. |
| [11] | ZHAO Yan-xia, ZHANG Jing-ying, SUN Jun-fei, WANG Jiang-hui, SUN Jia-bo, LV Xiao-hui. Analyses of Transcription and Metabolic Differential in the Flower Development Processes of ‘Rose rugosa cv. Plena’ [J]. Biotechnology Bulletin, 2023, 39(3): 184-195. |
| [12] | ZHAO Meng-liang, GUO Yi-ting, REN Yan-jing. Identification and Analysis of WRKY Transcription Factor Family Genes in Helianthus tuberosus [J]. Biotechnology Bulletin, 2023, 39(2): 116-125. |
| [13] | YU Bo, QIN Xiao-hui, ZHAO Yang. Mechanisms of Plant Sensing Drought Signals [J]. Biotechnology Bulletin, 2023, 39(11): 6-17. |
| [14] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| [15] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
