








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (3): 194-202.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0465
Previous Articles Next Articles
WANG Xiao-qin1( ), HUANG Yin-ping1, WANG Wei-qian2, WU Ping2, QUAN Shu1(
), HUANG Yin-ping1, WANG Wei-qian2, WU Ping2, QUAN Shu1( )
)
Received:2021-04-09
Online:2022-03-26
Published:2022-04-06
Contact:
QUAN Shu
E-mail:wxq870718522@163.com;shuquan@ecust.edu.cn
WANG Xiao-qin, HUANG Yin-ping, WANG Wei-qian, WU Ping, QUAN Shu. Expression and Purification of the MLL3SET Protein with a Site-directed Mutation of an Unnatural Amino Acid[J]. Biotechnology Bulletin, 2022, 38(3): 194-202.
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| Kan-F | TTCGCGTTCGCGTAAGTGTAGGCTGGAGCTGC |
| Kan-R | TTACGCGAAATACGGGCAGACATGGCCTGCC- CGGTTATTAATATCCTCCTTAGTTCCTATTCC |
| T7p07-F | GGAATTGTGAGCGGATAACAATTTCACACAGG- AAACAGCTATGAACACGATTAACATCGCTAAG |
| T7p07-R | CTCCAGCCTACACTTACGCGAACGCGAAGTCC |
| (T7p07+Kan)-F | GGAATTGTGAGCGGATAAC |
| (T7p07+Kan)-R | TTACGCGAAATACGGGCAG |
| F1 | CCAGAAAGGAGAAGAGCTCTCCTATGACTAT- AAGTTTGAC |
| R1 | GTCAAACTTA TAGTCATAG GAGAGCTCTTCT- CCTTTCTGG |
| F2 | GAGAAGCTTTATGAGTGTCAGAACCGTGGTG- TGTAC |
| R2 | GTACACACCACGGTTCTGACACTCATAAAGC- TTCTC |
| F3 | GTGGAGCTGTGTAGTGCCGGAAGTGGATG |
| R3 | CATCCACTTCCGGCACTACACAGCTCCAC |
Table 1 Primer sequences of PCR reactions
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| Kan-F | TTCGCGTTCGCGTAAGTGTAGGCTGGAGCTGC |
| Kan-R | TTACGCGAAATACGGGCAGACATGGCCTGCC- CGGTTATTAATATCCTCCTTAGTTCCTATTCC |
| T7p07-F | GGAATTGTGAGCGGATAACAATTTCACACAGG- AAACAGCTATGAACACGATTAACATCGCTAAG |
| T7p07-R | CTCCAGCCTACACTTACGCGAACGCGAAGTCC |
| (T7p07+Kan)-F | GGAATTGTGAGCGGATAAC |
| (T7p07+Kan)-R | TTACGCGAAATACGGGCAG |
| F1 | CCAGAAAGGAGAAGAGCTCTCCTATGACTAT- AAGTTTGAC |
| R1 | GTCAAACTTA TAGTCATAG GAGAGCTCTTCT- CCTTTCTGG |
| F2 | GAGAAGCTTTATGAGTGTCAGAACCGTGGTG- TGTAC |
| R2 | GTACACACCACGGTTCTGACACTCATAAAGC- TTCTC |
| F3 | GTGGAGCTGTGTAGTGCCGGAAGTGGATG |
| R3 | CATCCACTTCCGGCACTACACAGCTCCAC |
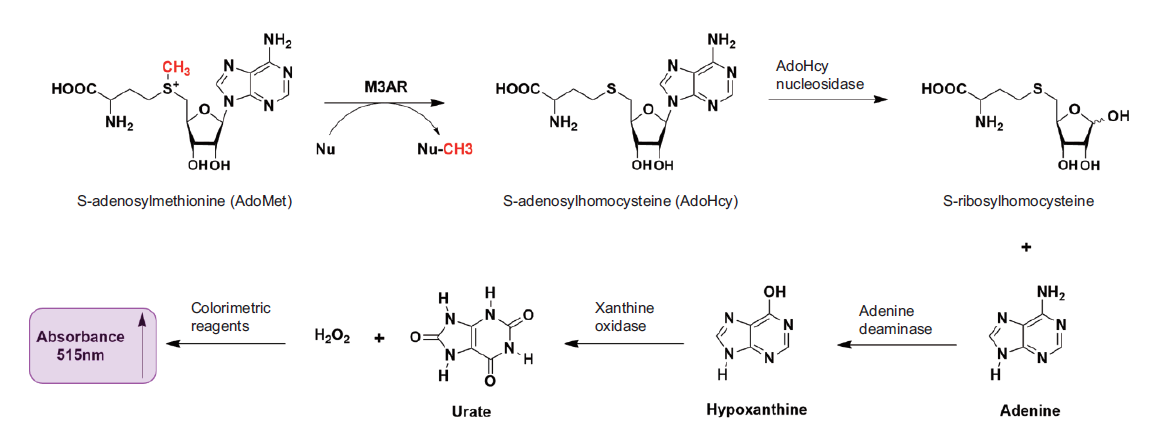
Fig. 1 Diagram of the multi-enzyme coupling reaction for determining MLL3SET* activity Under the catalysis of MLL3SET and AR compound(M3AR)and other enzymes,SAM undergoes multiple reactions. The final product N-(4-antipyryl)-3-chloro-5-sulfonate-ρ-benzoquinone-monoimine shows absorbance at 515 nm

Fig. 3 MLL3SET structural information(A)and the map of the expression plasmid (B) The cysteine at position 4 883 of MLL3SET is mutated to serine,serine at position 4 819 to cysteine,and asparagine at position 4 905 to the amber codon. A:Structural information of 6 cysteine residues in MLL3SET(PDB:5F6K). MLL3SET is colored in yellow;the substrate H3 is colored in aquamarine. The product,SAH,6 cysteines,and the two sites chosen for smFRET labeling are displayed as the stick model. The zinc ion is shown in the grey sphere. B:pET28b-His-sumo-mll3set C4883S S4819C N4905X plasmid map

Fig. 4 Expression test of the pET28b-His-sumo-mll3set C4883S plasmid M:Protein marker. U:Uninduced whole cell after lysis. I:Induced whole cell after lysis. S:Induced supernatant after lysis. P:Induced precipitation after lysis. 37℃:Expression at 37℃. 16℃:Expression at 16℃
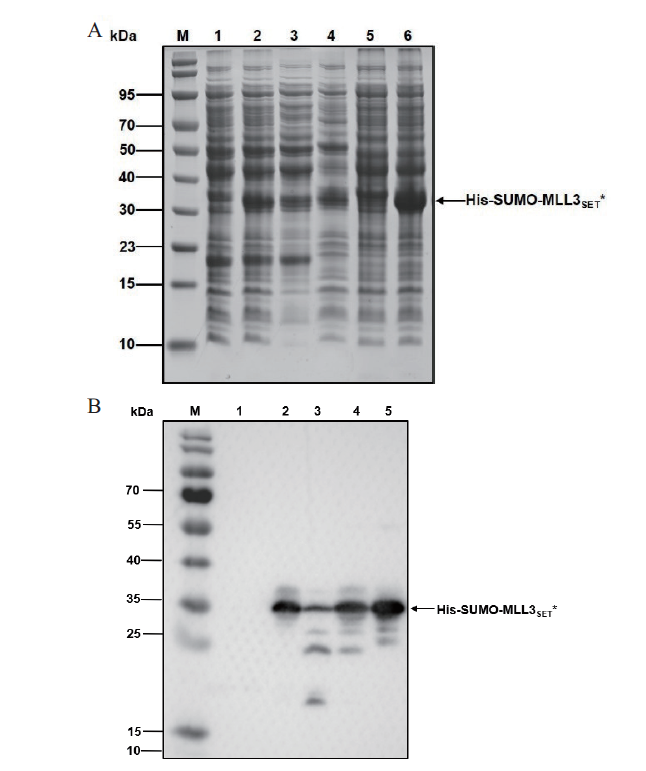
Fig. 5 Analysis of expressed His-SUMO-MLL3SET* A:SDS-PAGE analysis. B:Western blot analysis. M:Protein marker;1-4:The host strain was E. coli C321.ΔA. exp lacZ∷T7. 1:Uninduced whole cell after lysis. 2:Induced whole cell after lysis. 3:Supernatant of induced cells after lysis. 4:Precipitation of induced cells after lysis. 5-6:The host strain was E. coli BL21(DE3). 5:Uninduced whole cell after lysis. 6:Induced whole cell after lysis

Fig. 6 Analysis of purified His-SUMO-MLL3SET* M:Protein marker. 1:Induced whole cell. 2:Supernatant of the induced cells. 3:Precipitation of the induced cells. 4:Flow through before washing. 5:Flow through after washing with 50 mL buffer. 6:Flow through after washing with 1 L buffer. 7:Suspended Ni beads with buffer before imidazole elution. 8:Imidazole eluted sample. 9:Suspended Ni beads with buffer after imidazole elution

Fig. 9 Activity of His-SUMO-MLL3SET* measured by a multi-enzyme coupling reaction A:Comparison of the activity of M*AR and MAR. B:Standard curve of SAH concentration and A515
| [1] |
Shilatifard A. The COMPASS family of histone H3K4 methylases:mechanisms of regulation in development and disease pathogenesis[J]. Annu Rev Biochem, 2012, 81:65-95.
doi: 10.1146/annurev-biochem-051710-134100 pmid: 22663077 |
| [2] |
Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins[J]. Oncogene, 2001, 20(40):5695-5707.
pmid: 11607819 |
| [3] | 黎彦璟, 陈勇. 组蛋白甲基转移酶MLL1的结构与功能研究进展[J]. 中国细胞生物学学报, 2014, 36(7):857-868. |
| Li YJ, Chen Y. Progress in structural and functional studies of histone methyltransferase MLL1[J]. Chin J Cell Biol, 2014, 36(7):857-868. | |
| [4] |
Dehé PM, Dichtl B, Schaft D, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation[J]. J Biol Chem, 2006, 281(46):35404-35412.
doi: 10.1074/jbc.M603099200 URL |
| [5] |
Avdic V, Zhang P, Lanouette S, et al. Structural and biochemical insights into MLL1 core complex assembly[J]. Structure, 2011, 19(1):101-108.
doi: 10.1016/j.str.2010.09.022 URL |
| [6] |
Zhang Y, Mittal A, Reid J, et al. Evolving catalytic properties of the MLL family SET domain[J]. Structure, 2015, 23(10):1921-1933.
doi: S0969-2126(15)00324-X pmid: 26320581 |
| [7] |
Southall SM, Wong PS, Odho Z, et al. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks[J]. Mol Cell, 2009, 33(2):181-191.
doi: 10.1016/j.molcel.2008.12.029 pmid: 19187761 |
| [8] |
Li Y, Han J, Zhang Y, et al. Structural basis for activity regulation of MLL family methyltransferases[J]. Nature, 2016, 530(7591):447-452.
doi: 10.1038/nature16952 URL |
| [9] |
Sasmal DK, Pulido LE, Kasal S, et al. Single-molecule fluorescence resonance energy transfer in molecular biology[J]. Nanoscale, 2016, 8(48):19928-19944.
doi: 10.1039/C6NR06794H URL |
| [10] | Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET[J]. Nat Methods, 2008, 5(6):507-516. |
| [11] |
Zhang J, Fu Y, Lakowicz JR. Enhanced förster resonance energy transfer(FRET)on single metal particle[J]. J Phys Chem C Nanomater Interfaces, 2007, 111(1):50-56.
doi: 10.1021/jp062665e URL |
| [12] |
Zhang J, Fu Y, Chowdhury MH, et al. Enhanced förster resonance energy transfer on single metal particle. 2. dependence on donor-acceptor separation distance, particle size, and distance from metal surface[J]. J Phys Chem C, 2007, 111(32):11784-11792.
doi: 10.1021/jp067887r pmid: 19890406 |
| [13] |
Lee TC, Kang M, Kim CH, et al. Dual unnatural amino acid incorporation and click-chemistry labeling to enable single-molecule FRET studies of p97 folding[J]. Chembiochem, 2016, 17(11):981-984.
doi: 10.1002/cbic.v17.11 URL |
| [14] |
Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins[J]. Chem Rev, 2014, 114(9):4764-4806.
doi: 10.1021/cr400355w URL |
| [15] | Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli[J]. Nat Methods, 2006, 3(4):263-265. |
| [16] |
Young TS, Ahmad I, Yin JA, et al. An enhanced system for unnatural amino acid mutagenesis in E. coli[J]. J Mol Biol, 2010, 395(2):361-374.
doi: 10.1016/j.jmb.2009.10.030 URL |
| [17] |
Smolskaya S, Andreev Y. Site-specific incorporation of unnatural amino acids into Escherichia coli recombinant protein:methodology development and recent achievement[J]. Biomolecules, 2019, 9(7):255.
doi: 10.3390/biom9070255 URL |
| [18] |
Lin X, Yu ACS, Chan TF. Efforts and challenges in engineering the genetic code[J]. Life, 2017, 7(1):12.
doi: 10.3390/life7010012 URL |
| [19] |
Young DD, Schultz PG. Playing with the molecules of life[J]. ACS Chem Biol, 2018, 13(4):854-870.
doi: 10.1021/acschembio.7b00974 URL |
| [20] |
Dorgan KM, Wooderchak WL, Wynn DP, et al. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases[J]. Anal Biochem, 2006, 350(2):249-255.
doi: 10.1016/j.ab.2006.01.004 URL |
| [21] | Liu ZP. Histone methylation in heart development and cardiovascular disease[J]. Cardiac and Vascular Biology, 2016, 1:125-146. |
| [22] |
Bale TL, Baram TZ, Brown AS, et al. Early life programming and neurodevelopmental disorders[J]. Biol Psychiatry, 2010, 68(4):314-319.
doi: 10.1016/j.biopsych.2010.05.028 URL |
| [23] |
Millan MJ. An epigenetic framework for neurodevelopmental disorders:from pathogenesis to potential therapy[J]. Neuropharmacology, 2013, 68:2-82.
doi: 10.1016/j.neuropharm.2012.11.015 pmid: 23246909 |
| [24] |
Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders[J]. Nat Rev Genet, 2016, 17(1):9-18.
doi: 10.1038/nrg3999 URL |
| [1] | ZHAO Zhong-juan, YANG Kai, HU Jin-dong, WEI Yan-li, LI Ling, XU Wei-sheng, LI Ji-shun. Effects of Trichoderma harzianum ST02 on the Growth of Peppermint and Physicochemical Properties of Root Zone Soil Under Salt Stress [J]. Biotechnology Bulletin, 2022, 38(7): 224-235. |
| [2] | YUAN Cun-xia, LI Yan-nan, ZHANG Xiao-chong, YANG Rui, LIU Jian-li, LI Jing-yu. Physiological and Biochemical Response Characteristics of Bacillus sp. ZJS3 Under As3+ Stress [J]. Biotechnology Bulletin, 2022, 38(7): 236-246. |
| [3] | JIA Hai-hong, LI Bing-qing. Research Progress in the Post-translational Modification of Superoxide Dismutase [J]. Biotechnology Bulletin, 2022, 38(2): 237-244. |
| [4] | WU Qi-man, TIAN Shi-han, LI Yun-ye, PAN Ying-jie, ZHANG Ying. Effects of Microbial Fertilizer on Cucumis sativus L. Growth,Yield and Quality [J]. Biotechnology Bulletin, 2022, 38(1): 125-131. |
| [5] | CAO Ru-fei, LI Ze-xuan, XU Huan, ZHANG Sha, ZHANG Min-min, DAI Feng, DUAN Xiao-lei. Expression,Purification,and Crystallization of Pif1 Helicase from Bacteroides fragilis [J]. Biotechnology Bulletin, 2021, 37(9): 180-190. |
| [6] | YUAN Yuan, WANG Lei, SHI Ya-wei. Research Advances in Strategies for Improving the Activity of Microbial-derived Alkaline Proteases [J]. Biotechnology Bulletin, 2021, 37(5): 231-236. |
| [7] | CHEN Xiao-yu, ZHANG Jian, ZHANG Xin-ya, TANG Yu-ting, SHAO Yu-chen, LUO Zhi-dan, LU Chen. A Rapid and Accurate Method for Tth DNA Polymerase Activity Assay [J]. Biotechnology Bulletin, 2021, 37(5): 281-286. |
| [8] | XIE Guo-zhen, TANG Yuan, WU Yi, HUANG Li-li, TAN Zhou-jin. Effects of Total Glycosides of Qiwei Baizhu Powder on Intestinal Microbiota and Enzyme Activities in Diarrhea Mice [J]. Biotechnology Bulletin, 2021, 37(12): 124-131. |
| [9] | TAO Zhi-dong, HE Yan-hui, DENG Zi-he, SUN Lin-lin, WU Zhan-sheng. Screening of High-efficiency Cellulose-degrading Microorganism from Spent Lentinula edodes Substrate and Optimization of Its Enzyme Production [J]. Biotechnology Bulletin, 2021, 37(11): 158-165. |
| [10] | TIAN Geng, GAO Wei-qiang, CHEN Xiao-bo, ZHANG Chun-xiao. Directed Mutagenesis of β-mannanase Gene from Bacillus licheniformis KD-1 for Improving Enzyme Activity and Stability [J]. Biotechnology Bulletin, 2021, 37(10): 100-109. |
| [11] | WANG Xiang-feng, WANG Qiao, YUAN Hui-jun, WANG Li. Screening and Identification of High-yield Feruloyl Esterase Strains and Optimizing of the Enzyme Activity Assay Conditions [J]. Biotechnology Bulletin, 2020, 36(10): 135-141. |
| [12] | MENG Jian-yu, JI Jin-hua, JIA Li-juan, GUO Hui-qin, TAO Yu, FENG Fu-ying. Isolation of Cold-adapted Cellulose-degrading Bacteria Using Three Different Carbon Sources and Analysis on the Degrading Ability of Consortia [J]. Biotechnology Bulletin, 2019, 35(8): 77-84. |
| [13] | GENG Xiu-xiu, ZHOU Zheng-fu, LIU Ying-ying, PING Shu-zhen, WANG Jin. Cloning and Identification of Keratinase Gene from Deinococcus gobiensis I-0 [J]. Biotechnology Bulletin, 2019, 35(3): 65-70. |
| [14] | CHEN Jian-jun, LIU Liang-tao, CAO Xiang-lin. Cloning,Expression and Enzyme Production of Laccase Gene lac1680 in Phanerochaete chrysosporium [J]. Biotechnology Bulletin, 2018, 34(4): 214-220. |
| [15] | LIU Yan-xia,LI Shao-jie,FAN Zhen-chuan. Prokaryotic Expression,Purification and Polyclonal Antibody Preparation of Neurospora crassa ERG-11 Protein Antigen [J]. Biotechnology Bulletin, 2017, 33(9): 216-222. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||