








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (12): 149-155.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0400
Previous Articles Next Articles
Received:2022-05-23
Online:2022-12-26
Published:2022-12-29
Contact:
LIU Yong-zhe
E-mail:chenduozx2751@163.com;huntertry@163.com
CHEN Duo, LIU Yong-zhe. Prokaryotic Expression,Purification and Crystallization of N-terminal Domain of Nucleocapsid Protein in SARS-CoV-2[J]. Biotechnology Bulletin, 2022, 38(12): 149-155.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| N-N-F | GGAATTCCATATGAATAATACTGCGTCTTGGTT |
| N-N-R | CCGCTCGAGTTACCCTTCTGCGTAGAAGCCTT |
Table 1 Primer sequences
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| N-N-F | GGAATTCCATATGAATAATACTGCGTCTTGGTT |
| N-N-R | CCGCTCGAGTTACCCTTCTGCGTAGAAGCCTT |
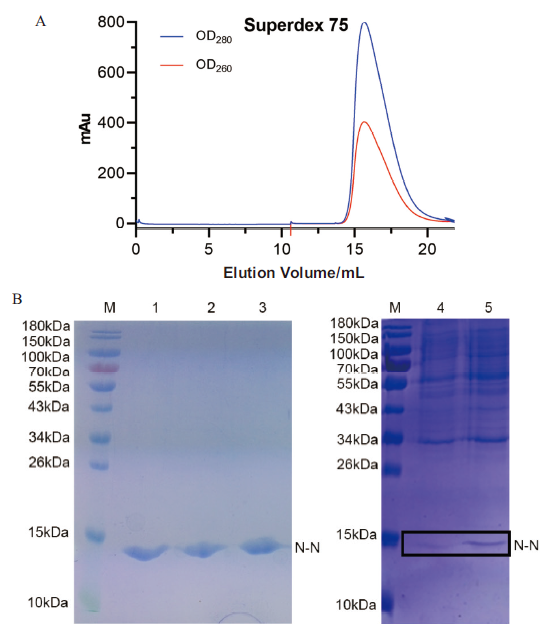
Fig. 3 Expression and purification of N-N protein A:The N-N protein samples were purified by Superdex 75 column. B:The purified N-N protein samples were analyzed on SDS-PAGE. M:Protein marker. 1-3:The N-N protein;4:Before-induced.5:After-induced
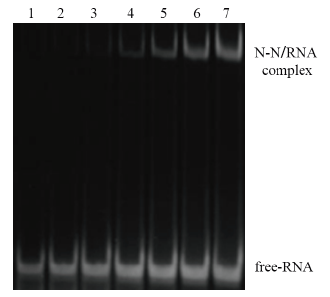
Fig. 4 EMSA result of binding of N-N protein and RNA 1:Normal RNA control. 2-7:Incubation of RNA and N-N protein(The N-N protein concentrations correspond to 1,2,3,4,5 and 6 mg/mL respectively)
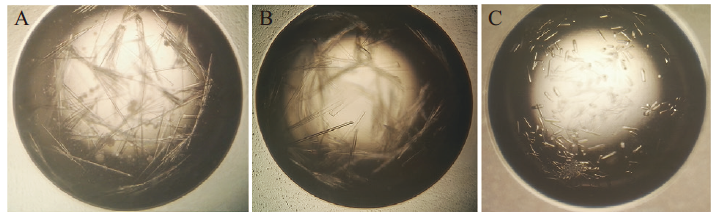
Fig. 5 Optimization of N protein crystal A:Making Initial crystal by PEG/Ion Screen 23. B:Making further crystal by adding Crystal Screen 45. C:Making final crystal by adding Additive Screen 25
| [1] |
Dong ES, Du HR, Gardner L. An interactive web-based dashboard to track COVID-19 in real time[J]. Lancet Infect Dis, 2020, 20(5):533-534.
doi: S1473-3099(20)30120-1 pmid: 32087114 |
| [2] |
Huang CL, Wang YM, Li XW, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China[J]. Lancet, 2020, 395(10223):497-506.
doi: S0140-6736(20)30183-5 pmid: 31986264 |
| [3] |
Anand KB, Karade S, Sen S, et al. SARS-CoV-2:camazotz’s curse[J]. Med J Armed Forces India, 2020, 76(2):136-141.
doi: 10.1016/j.mjafi.2020.04.008 URL |
| [4] |
Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing[J]. Nature, 2020, 583(7816):459-468.
doi: 10.1038/s41586-020-2286-9 URL |
| [5] |
Romano M, Ruggiero A, Squeglia F, et al. A structural view of SARS-CoV-2 RNA replication machinery:RNA synthesis, proofreading and final capping[J]. Cells, 2020, 9(5):1267.
doi: 10.3390/cells9051267 URL |
| [6] |
Hu B, Guo H, Zhou P, et al. Characteristics of SARS-CoV-2 and COVID-19[J]. Nat Rev Microbiol, 2021, 19(3):141-154.
doi: 10.1038/s41579-020-00459-7 pmid: 33024307 |
| [7] |
Yang D, Leibowitz JL. The structure and functions of coronavirus genomic 3' and 5' ends[J]. Virus Res, 2015, 206:120-133.
doi: 10.1016/j.virusres.2015.02.025 pmid: 25736566 |
| [8] |
Uddin M, Mustafa F, Rizvi TA, et al. SARS-CoV-2/COVID-19:viral genomics, epidemiology, vaccines, and therapeutic interventions[J]. Viruses, 2020, 12(5):526.
doi: 10.3390/v12050526 URL |
| [9] |
Yao HP, Song YT, Chen Y, et al. Molecular architecture of the SARS-CoV-2 virus[J]. Cell, 2020, 183(3):730-738. e13.
doi: 10.1016/j.cell.2020.09.018 pmid: 32979942 |
| [10] |
Ye QZ, West AMV, Silletti S, et al. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein[J]. Protein Sci, 2020, 29(9):1890-1901.
doi: 10.1002/pro.3909 URL |
| [11] | Peng Y, Du N, Lei YQ, et al. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design[J]. EMBO J, 2020, 39(20):e105938. |
| [12] | Jia Z, Liu C, Chen Y, et al. Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies[J]. FEBS J, 2021. Doi:10.1111/febs.16239. |
| [13] |
张西西, 张怡青, 李玉林, 等. 新型冠状病毒(SARS-CoV-2)N蛋白C端重组蛋白的原核表达、纯化及应用[J]. 生物技术通报, 2021, 37(5):92-97.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0902 URL |
| Zhang XX, Zhang YQ, Li YL, et al. Prokaryotic expression, purification and application of N protein C-terminal recombinant protein in novel coronavirus(SARS-CoV-2)[J]. Biotechnol Bull, 2021, 37(5):92-97. | |
| [14] |
Wu C, Qavi AJ, Moyle AB, et al. Domain-specific biochemical and serological characterization of SARS-CoV-2 nucleocapsid protein[J]. STAR Protoc, 2021, 2(4):100906.
doi: 10.1016/j.xpro.2021.100906 URL |
| [15] |
Zeng WH, Liu GF, Ma H, et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein[J]. Biochem Biophys Res Commun, 2020, 527(3):618-623.
doi: 10.1016/j.bbrc.2020.04.136 URL |
| [16] |
Gupta Y, Kumar S, Zak SE, et al. Antiviral evaluation of hydroxyethylamine analogs:Inhibitors of SARS-CoV-2 main protease(3CLpro), a virtual screening and simulation approach[J]. Bioorg Med Chem, 2021, 47:116393.
doi: 10.1016/j.bmc.2021.116393 URL |
| [17] |
Samper IC, McMahon CJ, Schenkel MS, et al. Electrochemical immunoassay for the detection of SARS-CoV-2 nucleocapsid protein in nasopharyngeal samples[J]. Anal Chem, 2022, 94(11):4712-4719.
doi: 10.1021/acs.analchem.1c04966 pmid: 35263100 |
| [18] |
Low JS, Vaqueirinho D, Mele F, et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2[J]. Science, 2021, 372(6548):1336-1341.
doi: 10.1126/science.abg8985 pmid: 34006597 |
| [19] |
López-Muñoz AD, Kosik I, Holly J, et al. Cell surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity[J]. bioRxiv, 2021. DOI:10.1101/2021.12.10.472169.
doi: 10.1101/2021.12.10.472169 URL |
| [20] |
Xie CZ, Ding HJ, Ding JZ, et al. Preparation of highly specific monoclonal antibodies against SARS-CoV-2 nucleocapsid protein and the preliminary development of antigen detection test strips[J]. J Med Virol, 2022, 94(4):1633-1640.
doi: 10.1002/jmv.27520 URL |
| [21] | Li DD, Li JM. Immunologic testing for SARS-CoV-2 infection from the antigen perspective[J]. J Clin Microbiol, 2021, 59(5):e02160-e02120. |
| [22] |
Wang YT, Long XY, Ding X, et al. Novel nucleocapsid protein-targeting phenanthridine inhibitors of SARS-CoV-2[J]. Eur J Med Chem, 2022, 227:113966.
doi: 10.1016/j.ejmech.2021.113966 URL |
| [23] |
Lucas C, Vogels CBF, Yildirim I, et al. Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity[J]. Nature, 2021, 600(7889):523-529.
doi: 10.1038/s41586-021-04085-y URL |
| [24] |
Pitcovski J, Gruzdev N, Abzach A, et al. Oral subunit SARS-CoV-2 vaccine induces systemic neutralizing IgG, IgA and cellular immune responses and can boost neutralizing antibody responses primed by an injected vaccine[J]. Vaccine, 2022, 40(8):1098-1107.
doi: 10.1016/j.vaccine.2022.01.025 URL |
| [25] |
Chatzileontiadou DSM, Szeto C, Jayasinghe D, et al. Protein purification and crystallization of HLA-A 02:01 in complex with SARS-CoV-2 peptides[J]. STAR Protoc, 2021, 2(3):100635.
doi: 10.1016/j.xpro.2021.100635 URL |
| [26] |
Cubuk J, Alston JJ, Incicco JJ, et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA[J]. Nat Commun, 2021, 12(1):1936.
doi: 10.1038/s41467-021-21953-3 pmid: 33782395 |
| [27] |
Savastano A, Ibáñez de Opakua A, Rankovic M, et al. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates[J]. Nat Commun, 2020, 11(1):6041.
doi: 10.1038/s41467-020-19843-1 pmid: 33247108 |
| [28] |
Jack A, Ferro LS, Trnka MJ, et al. SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA[J]. PLoS Biol, 2021, 19(10):e3001425.
doi: 10.1371/journal.pbio.3001425 URL |
| [29] |
Parashar NC, Poddar J, Chakrabarti S, et al. Repurposing of SARS-CoV nucleocapsid protein specific nuclease resistant RNA aptamer for therapeutics against SARS-CoV-2[J]. Infect Genet Evol, 2020, 85:104497.
doi: 10.1016/j.meegid.2020.104497 URL |
| [30] | Tatar G, Ozyurt E, Turhan K. Computational drug repurposing study of the RNA binding domain of SARS-CoV-2 nucleocapsid protein with antiviral agents[J]. Biotechnol Prog, 2021, 37(2):e3110. |
| [1] | LI Wen-shuo, WANG Lin-song, DU Gui-cai, GUO Qun-qun, ZHANG Ting-ting, YANG Hong LI Rong-gui. Gene Cloning of an Aldehyde Dehydrogenase from Bursaphelenchus xylophilus and Biochemical Characterization [J]. Biotechnology Bulletin, 2022, 38(9): 207-214. |
| [2] | QIN Xue-jing, WANG Yu-han, CAO Yi-bo, ZHANG Ling-yun. Prokaryotic Expression and Preparation of Polyclonal Antibody of PwHAP5 Gene in Picea wilsonii [J]. Biotechnology Bulletin, 2022, 38(8): 142-149. |
| [3] | YI Fang, LAI Peng-cheng, ZHENG Xi-ao, HU Shuai, GAO Yan-li. Research on the Preparation and Purification of Kod DNA Polymerase [J]. Biotechnology Bulletin, 2022, 38(5): 183-190. |
| [4] | WANG Jia-li, HE Si-qi, KANG Zi-xi, WANG Jian-xun. Antibody Phage Display Technology and Its Application in the Discovery of Anti-SARS-CoV-2 Antibodies [J]. Biotechnology Bulletin, 2022, 38(5): 248-256. |
| [5] | WANG Qiao-ju, HU Yu-meng, WEN Ya-ya, SONG Li, MENG Chuang, PAN Zhi-ming, JIAO Xin-an. Expression and Activity Identification of SARS-CoV-2 S1 Protein [J]. Biotechnology Bulletin, 2022, 38(3): 157-163. |
| [6] | LIU Xiao-mei, WANG Dong-xin, ZHANG Chun, WEI Shuang-shi. Inhibition of AAV-mediated RNAi to SARS-CoV-2 S Gene Expression [J]. Biotechnology Bulletin, 2022, 38(3): 188-193. |
| [7] | WANG Xiao-qin, HUANG Yin-ping, WANG Wei-qian, WU Ping, QUAN Shu. Expression and Purification of the MLL3SET Protein with a Site-directed Mutation of an Unnatural Amino Acid [J]. Biotechnology Bulletin, 2022, 38(3): 194-202. |
| [8] | ZHANG Feng-wen, ZHOU Li-ya, DONG Chao, SHI Yan-mao. Purification of Antioxidant Peptides from Natto Supernatant and Study on Its Activity [J]. Biotechnology Bulletin, 2022, 38(2): 158-165. |
| [9] | CAO Ru-fei, LI Ze-xuan, XU Huan, ZHANG Sha, ZHANG Min-min, DAI Feng, DUAN Xiao-lei. Expression,Purification,and Crystallization of Pif1 Helicase from Bacteroides fragilis [J]. Biotechnology Bulletin, 2021, 37(9): 180-190. |
| [10] | TIAN Jia-hui, FENG Jia-li, LU Jun-hua, MAO Lin-jing, HU Zhu-ran, WANG Ying, CHU Jie. Isolation,Purification and Characterization of Laccase LacT-1 from Cerrena unicolor [J]. Biotechnology Bulletin, 2021, 37(8): 186-194. |
| [11] | ZHANG Xi-xi, ZHANG Yi-qing, LI Yu-lin, HAN Xiao, WANG Guo-qiang, WANG Xiao-jun, WANG Xu-dong, WANG Yun-long. Prokaryotic Expression,Purification and Application of N Protein C-terminal Recombinant Protein in Novel Coronavirus(SARS-CoV-2) [J]. Biotechnology Bulletin, 2021, 37(5): 92-97. |
| [12] | LI Jia-jun, ZHENG Xiao, SHENG Jie, XU Yao. Novel Coronavirus and Research Progress of Related Clinical Detection Methods [J]. Biotechnology Bulletin, 2021, 37(4): 282-292. |
| [13] | WANG Zhi-xin, LU Lei-zhen, ZHOU Jing-bo, FENG Cheng-ling, JIA Zi-wei, NING Ya-wei, JIA Ying-min. Advance in the Research of the Antifungal Peptides [J]. Biotechnology Bulletin, 2021, 37(3): 206-218. |
| [14] | GAO Jing-xi, GAO Ke-xing, LU Fei, JI Feng, GUO Zhi-gang. Prediction of SARS-CoV-2 S Protein B Cell Antigenic Epitope Cross-immunizing with SARS-CoV [J]. Biotechnology Bulletin, 2021, 37(10): 169-178. |
| [15] | TANG Lu, DONG Li-ping, YIN Mo-li, LIU Lei, DONG Yuan, WANG Hui-yan. Preparation and Identification of a Novel FGF20 Monoclonal Antibody [J]. Biotechnology Bulletin, 2021, 37(10): 179-185. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
