








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (1): 187-198.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1033
Previous Articles Next Articles
DUAN Min-jie1( ), LI Yi-fei1, YANG Xiao-miao1, WANG Chun-ping2, HUANG Qi-zhong1, HUANG Ren-zhong1, ZHANG Shi-cai1(
), LI Yi-fei1, YANG Xiao-miao1, WANG Chun-ping2, HUANG Qi-zhong1, HUANG Ren-zhong1, ZHANG Shi-cai1( )
)
Received:2022-08-23
Online:2023-01-26
Published:2023-02-02
Contact:
ZHANG Shi-cai
E-mail:swemart@163.com;shicaiz@126.com
DUAN Min-jie, LI Yi-fei, YANG Xiao-miao, WANG Chun-ping, HUANG Qi-zhong, HUANG Ren-zhong, ZHANG Shi-cai. Identification of Zinc Finger Protein DnaJ-Like Gene Family in Capsicum annuum and Its Expression Analysis Responses to High Temperature Stress[J]. Biotechnology Bulletin, 2023, 39(1): 187-198.
| 基因名称 Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| CaDNAJE2 | TACCCCGAAAAGTGTCCGC | TCGTCGAAAACCACCCTATGAG |
| CaDNAJE3 | TTCTGCTCTATGGGGCTCTTAC | TTTTCCTCCACGGGTTCCT |
| CaDNAJE11 | AACACTCAGCCCTGCTTCCC | GCTGAATGCCACTCCCTTGAC |
| CaDNAJE13 | TGGTGGTGCCGATGAGATT | GCATTCGTCGCAGAGGATAA |
| CaDNAJE15 | CACCAAAGTGAGGAGCATCG | AATCCACCAAGAAATCCAGCA |
| CaDNAJE17 | ATTCCAATCCCAGCCACAA | CAACAGCAGCGATGACCAA |
| CaDNAJE19 | GCTCAGCAAGCGGTAAAT | TGCCATCACCATCCCAGT |
| CaDNAJE21 | CCCTTTCACTTCCAACCCC | TCAGTCCCATTTAGCCCATTTA |
| CaDNAJE23 | CGGTGCCGCTCTGGTGAAATC | CGACAACCCACGTCCCCATCT |
| CaDNAJE24 | TGGGAAGAGTCTATTGATGAACG | ATTGGTAGACACGCTTGGTTTT |
| CaDNAJE27 | ACGCAAAGTAAGAAGAAAGCAGC | CGAGAACAGTATCCAAGCCCAC |
| CaHsfB5 | AAAGTTCCAGAAAGGGTGC | TCCATACATTGAGTGAGGC |
| CaHSP22.0 | CCCCACTCACCATCCCAAAA | CCCCACTGATTCTCAACACCCTA |
| CaHSP16.4 | ATGTCAAAGATGATCAGCTTATTG | GGGTACTCAACACGGGACAC |
| CaUBI3 | GGACCCAAACCCCGATGAT | CACCCCAAGCACAATAAGACA |
Table 1 Primers used for quantitative real-time PCR
| 基因名称 Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| CaDNAJE2 | TACCCCGAAAAGTGTCCGC | TCGTCGAAAACCACCCTATGAG |
| CaDNAJE3 | TTCTGCTCTATGGGGCTCTTAC | TTTTCCTCCACGGGTTCCT |
| CaDNAJE11 | AACACTCAGCCCTGCTTCCC | GCTGAATGCCACTCCCTTGAC |
| CaDNAJE13 | TGGTGGTGCCGATGAGATT | GCATTCGTCGCAGAGGATAA |
| CaDNAJE15 | CACCAAAGTGAGGAGCATCG | AATCCACCAAGAAATCCAGCA |
| CaDNAJE17 | ATTCCAATCCCAGCCACAA | CAACAGCAGCGATGACCAA |
| CaDNAJE19 | GCTCAGCAAGCGGTAAAT | TGCCATCACCATCCCAGT |
| CaDNAJE21 | CCCTTTCACTTCCAACCCC | TCAGTCCCATTTAGCCCATTTA |
| CaDNAJE23 | CGGTGCCGCTCTGGTGAAATC | CGACAACCCACGTCCCCATCT |
| CaDNAJE24 | TGGGAAGAGTCTATTGATGAACG | ATTGGTAGACACGCTTGGTTTT |
| CaDNAJE27 | ACGCAAAGTAAGAAGAAAGCAGC | CGAGAACAGTATCCAAGCCCAC |
| CaHsfB5 | AAAGTTCCAGAAAGGGTGC | TCCATACATTGAGTGAGGC |
| CaHSP22.0 | CCCCACTCACCATCCCAAAA | CCCCACTGATTCTCAACACCCTA |
| CaHSP16.4 | ATGTCAAAGATGATCAGCTTATTG | GGGTACTCAACACGGGACAC |
| CaUBI3 | GGACCCAAACCCCGATGAT | CACCCCAAGCACAATAAGACA |
| 基序 Motif | 长度 Length/aa | 氨基酸保守基序 Conserved sequence of amino acid |
|---|---|---|
| motif 1 | 20 | CEGCGGSGFVPCSTCNGSGK |
| motif 2 | 33 | EDKIVIYFTSLRGIRKTFEDCNTVRMILESFRV |
| motif 3 | 45 | LQNVLEGKKVSLPRVFIKGRYIGGAEEIKQL- HEEGELKKLLEGJP |
| motif 4 | 15 | CPECNENGLIRCPIC |
| motif 5 | 15 | VDERDVSMDSGFREE |
Table 2 Conserved motif of the DNAJE proteins in pepper
| 基序 Motif | 长度 Length/aa | 氨基酸保守基序 Conserved sequence of amino acid |
|---|---|---|
| motif 1 | 20 | CEGCGGSGFVPCSTCNGSGK |
| motif 2 | 33 | EDKIVIYFTSLRGIRKTFEDCNTVRMILESFRV |
| motif 3 | 45 | LQNVLEGKKVSLPRVFIKGRYIGGAEEIKQL- HEEGELKKLLEGJP |
| motif 4 | 15 | CPECNENGLIRCPIC |
| motif 5 | 15 | VDERDVSMDSGFREE |
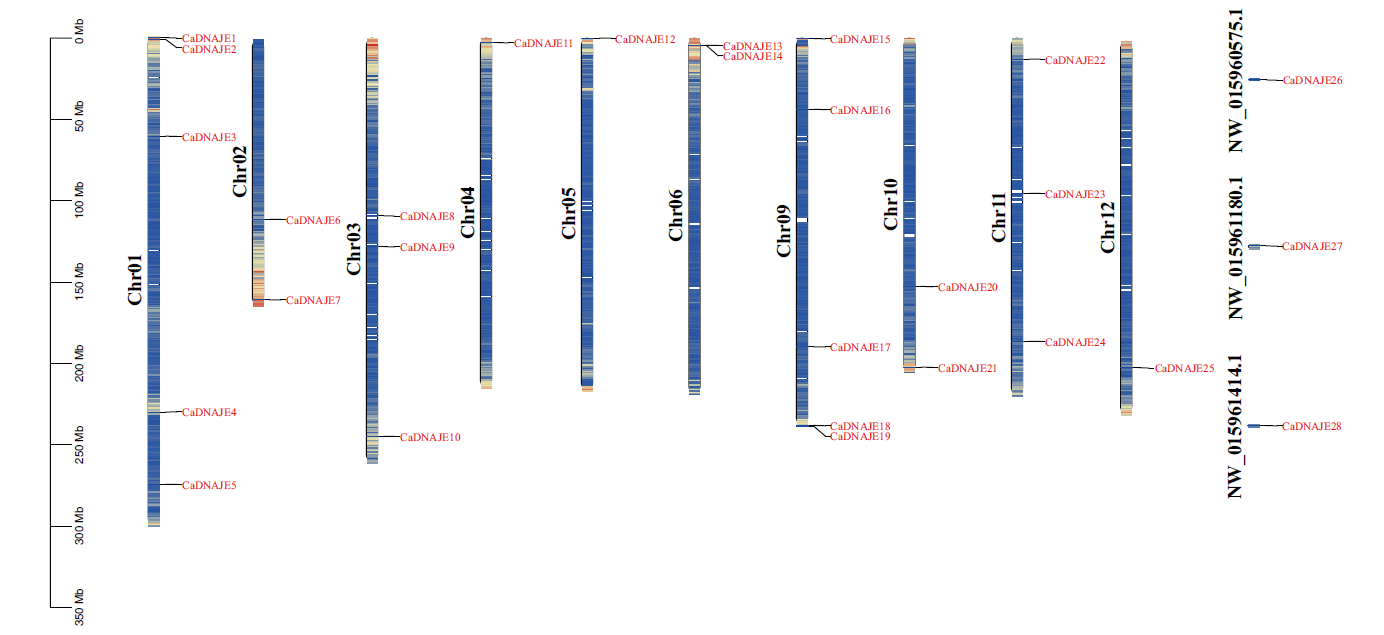
Fig. 1 Chromosomal location of DNAJE gene family members in pepper(Capsicum annuum) Chromosomal color ranging from green to red indicates the density of gene
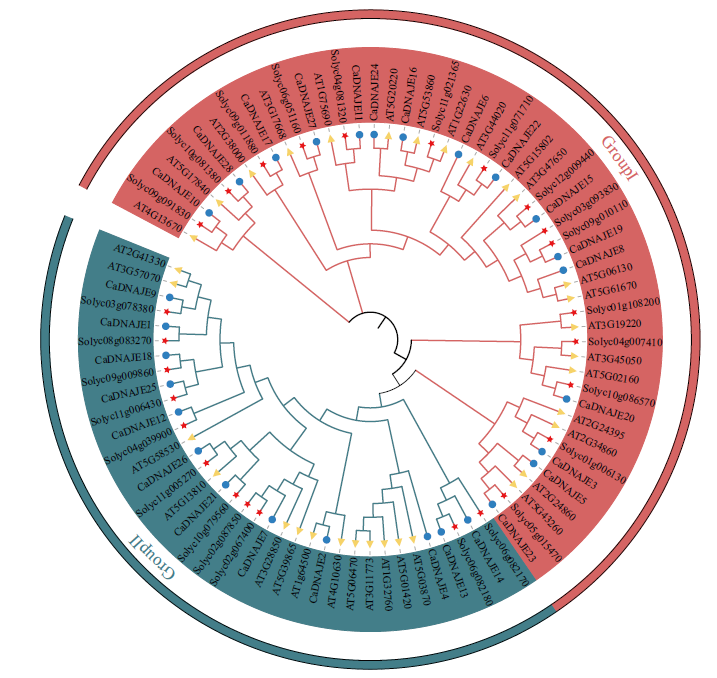
Fig. 2 Phylogenetic tree of DNAJE gene family members in pepper, Arabidopsis and tomato(Solanum lyc-opersicum) The blue solid dot indicates pepper DNAJE protein, yellow triangle indicates tomato DNAJE protein, and red star indicates Arabidopsis DNAJE protein
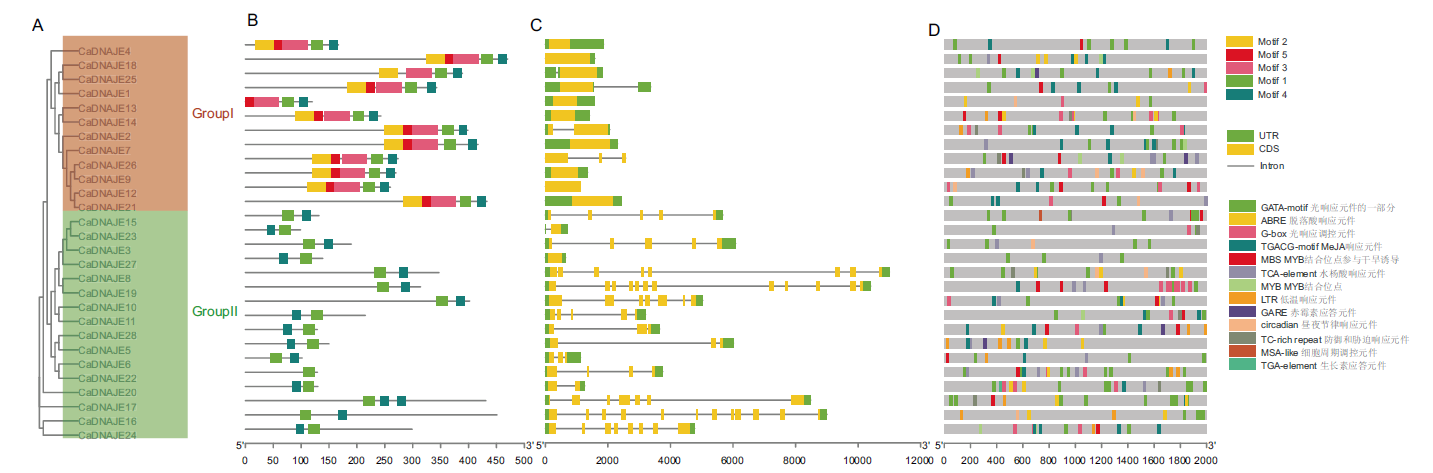
Fig. 3 Analysis of conserved motifs, gene structure and cis-acting elements of DNAJE gene family in pepper A: CaDNAJE protein evolution. B: Conserved motif. C: Gene structure. D: cis-acting element distribution
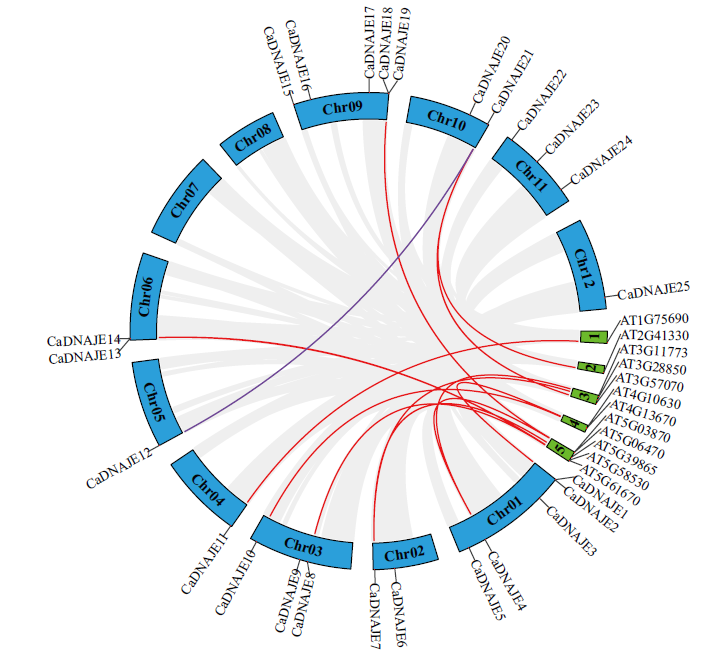
Fig. 4 Collinearity analysis of DNAJE gene family between pepper and Arabidopsis Chr and the number 1-5 indicate the chromosomes of pepper and Arabidopsis, respectively. The red lines indicate DNAJE collinearity between interspecific. The purple lines indicate DNAJE collinearity between intraspecific. The Gray lines indicate represent collinearity of all gene membres between interspecific
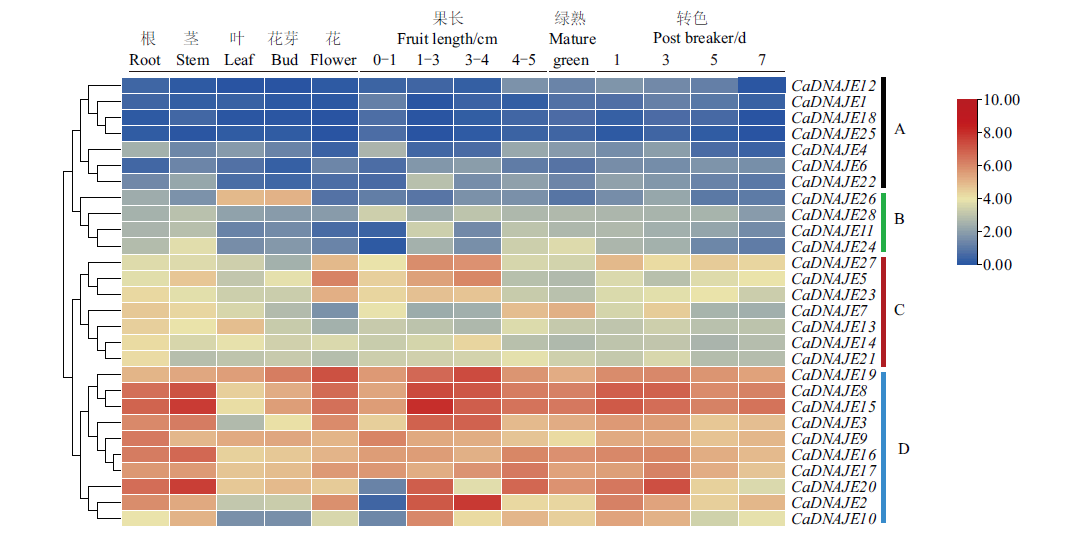
Fig. 5 Expression profile analysis of pepper DNAJE genes in different tissues and fruit development A, B, C and D indicate the clustering of different expression patterns

Fig. 6 Expression analysis of genes related to high temperature stress in pepper * indicates significant differences at 0.05 level, and ** indicates significant differences at 0.01 level
| [1] |
Georgopoulos CP, Lundquist-Heil A, Yochem J, et al. Identification of the E. coli dnaJ gene product[J]. Mol Gen Genet, 1980, 178(3): 583-588.
doi: 10.1007/BF00337864 URL |
| [2] |
Silver PA, Way JC. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity[J]. Cell, 1993, 74(1): 5-6.
pmid: 8334705 |
| [3] |
Caplan AJ, Cyr DM, Douglas MG. Eukaryotic homologues of Escherichia coli dnaJ: a diverse protein family that functions with hsp70 stress proteins[J]. Mol Biol Cell, 1993, 4(6): 555-563.
pmid: 8374166 |
| [4] |
Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog[J]. J Biol Chem, 1992, 267(29): 20927-20931.
pmid: 1400408 |
| [5] |
Walsh P, Bursac D, Law YC, et al. The J-protein family: modulating protein assembly, disassembly and translocation[J]. EMBO Rep, 2004, 5(6): 567-571.
doi: 10.1038/sj.embor.7400172 pmid: 15170475 |
| [6] |
Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity[J]. Nat Rev Mol Cell Biol, 2010, 11(8): 579-592.
doi: 10.1038/nrm2941 URL |
| [7] |
Ajit Tamadaddi C, Sahi C. J domain independent functions of J proteins[J]. Cell Stress Chaperones, 2016, 21(4): 563-570.
doi: 10.1007/s12192-016-0697-1 URL |
| [8] |
Pulido P, Leister D. Novel DNAJ-related proteins in Arabidopsis thaliana[J]. New Phytol, 2018, 217(2): 480-490.
doi: 10.1111/nph.14827 pmid: 29271039 |
| [9] |
Navrot N, Gelhaye E, Jacquot JP, et al. Identification of a new family of plant proteins loosely related to glutaredoxins with four CxxC motives[J]. Photosynth Res, 2006, 89(2/3): 71-79.
doi: 10.1007/s11120-006-9083-7 URL |
| [10] | 陈斯茗, 卢山. 植物DnaJ-Like锌指蛋白的功能研究[J]. 植物生理学报, 2015, 51(5): 586-592. |
|
Chen SM, Lu S. Recent studies on the DNA J-like zinc finger proteins[J]. Plant Physiol J, 2015, 51(5): 586-592.
doi: 10.1104/pp.51.3.586 URL |
|
| [11] |
Lu S, van Eck J, Zhou XJ, et al. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation[J]. Plant Cell, 2006, 18(12): 3594-3605.
doi: 10.1105/tpc.106.046417 pmid: 17172359 |
| [12] |
de Saint Germain A, Bonhomme S, Boyer FD, et al. Novel insights into strigolactone distribution and signalling[J]. Curr Opin Plant Biol, 2013, 16(5): 583-589.
doi: 10.1016/j.pbi.2013.06.007 pmid: 23830996 |
| [13] |
Amiya R, Shapira M. ZnJ6 is a thylakoid membrane DnaJ-like chaperone with oxidizing activity in Chlamydomonas reinhardtii[J]. Int J Mol Sci, 2021, 22(3): 1136.
doi: 10.3390/ijms22031136 URL |
| [14] |
Brutnell TP, Sawers RJ, Mant A, et al. BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize[J]. Plant Cell, 1999, 11(5): 849-864.
pmid: 10330470 |
| [15] |
Doron L, Segal N, Gibori H, et al. The BSD2 ortholog in Chlam-ydomonas reinhardtii is a polysome-associated chaperone that co-migrates on sucrose gradients with the rbcL transcript encoding the Rubisco large subunit[J]. Plant J, 2014, 80(2): 345-355.
doi: 10.1111/tpj.12638 URL |
| [16] |
Shimada H, Mochizuki M, Ogura K, et al. Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase[J]. Plant Cell, 2007, 19(10): 3157-3169.
doi: 10.1105/tpc.107.051714 pmid: 17921316 |
| [17] |
Tanz SK, Kilian J, Johnsson C, et al. The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings[J]. Plant J, 2012, 69(5): 743-754.
doi: 10.1111/j.1365-313X.2011.04833.x URL |
| [18] |
Muñoz-Nortes T, Pérez-Pérez JM, Ponce MR, et al. The ANGULA-TA7 gene encodes a DnaJ-like zinc finger-domain protein involved in chloroplast function and leaf development in Arabidopsis[J]. Plant J, 2017, 89(5): 870-884.
doi: 10.1111/tpj.13466 URL |
| [19] |
Albrecht V, Ingenfeld A, Apel K. Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves[J]. Plant Mol Biol, 2008, 66(6): 599-608.
doi: 10.1007/s11103-008-9291-y URL |
| [20] |
Zagari N, Sandoval-Ibañez O, Sandal N, et al. SNOWY COTYLEDON 2 promotes chloroplast development and has a role in leaf variegation in both Lotus japonicus and Arabidopsis thaliana[J]. Mol Plant, 2017, 10(5): 721-734.
doi: 10.1016/j.molp.2017.02.009 URL |
| [21] |
Fristedt R, Williams-Carrier R, Merchant SS, et al. A thylakoid membrane protein harboring a DnaJ-type zinc finger domain is required for photosystem I accumulation in plants[J]. J Biol Chem, 2014, 289(44): 30657-30667.
doi: 10.1074/jbc.M114.587758 pmid: 25228689 |
| [22] |
Lu Y, Hall DA, Last RL. A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana[J]. Plant Cell, 2011, 23(5): 1861-1875.
doi: 10.1105/tpc.111.085456 URL |
| [23] |
Whippo CW, Khurana P, Davis PA, et al. THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility[J]. Curr Biol, 2011, 21(1): 59-64.
doi: 10.1016/j.cub.2010.11.059 pmid: 21185188 |
| [24] |
Zhu JK. Abiotic stress signaling and responses in plants[J]. Cell, 2016, 167(2): 313-324.
doi: 10.1016/j.cell.2016.08.029 URL |
| [25] | 邹学校, 马艳青, 戴雄泽, 等. 辣椒在中国的传播与产业发展[J]. 园艺学报, 2020, 47(9): 1715-1726. |
| Zou XX, Ma YQ, Dai XZ, et al. Spread and industry development of pepper in China[J]. Acta Hortic Sin, 2020, 47(9): 1715-1726. | |
| [26] |
Qin C, Yu C, Shen Y, et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization[J]. PNAS, 2014, 111(14): 5135-5140.
doi: 10.1073/pnas.1400975111 URL |
| [27] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [28] |
Kim S, Park M, Yeom SI, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species[J]. Nat Genet, 2014, 46(3): 270-278.
doi: 10.1038/ng.2877 URL |
| [29] | 樊芳菲. 辣椒DnaJ基因家族:鉴定、结构特征及表达分析[D]. 广州: 华南农业大学, 2017. |
| Fan FF. The DnaJ gene family in pepper(Capsicum annuum L.): comprehensive identification, characterization and expression profiles[D]. Guangzhou: South China Agricultural University, 2017. | |
| [30] |
Wang J, Yang GB, Chen Y, et al. Genome-wide characterization and anthocyanin-related expression analysis of the B-BOX gene family in Capsicum annuum L[J]. Front Genet, 2022, 13: 847328.
doi: 10.3389/fgene.2022.847328 URL |
| [31] |
Arce-Rodríguez ML, Martínez O, Ochoa-Alejo N. Genome-wide identification and analysis of the MYB transcription factor gene family in chili pepper(Capsicum spp.)[J]. Int J Mol Sci, 2021, 22(5): 2229.
doi: 10.3390/ijms22052229 URL |
| [32] |
Jin JH, Wang M, Zhang HX, et al. Genome-wide identification of the AP2/ERF transcription factor family in pepper(Capsicum annuum L.)[J]. Genome, 2018, 61(9): 663-674.
doi: 10.1139/gen-2018-0036 URL |
| [33] |
Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40[J]. Cell Stress Chaperones, 2003, 8(4): 309-316.
doi: 10.1379/1466-1268(2003)008<0309:MFROHF>2.0.CO;2 URL |
| [34] |
Szabo A, Korszun R, Hartl FU, et al. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates[J]. EMBO J, 1996, 15(2): 408-417.
pmid: 8617216 |
| [35] |
Osorio CE. The role of orange gene in carotenoid accumulation: manipulating chromoplasts toward a colored future[J]. Front Plant Sci, 2019, 10: 1235.
doi: 10.3389/fpls.2019.01235 URL |
| [36] |
Zhou XJ, Welsch R, Yang Y, et al. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis[J]. Proc Natl Acad Sci USA, 2015, 112(11): 3558-3563.
doi: 10.1073/pnas.1420831112 URL |
| [37] |
Sun TH, Zhou F, Huang XQ, et al. ORANGE represses chloroplast biogenesis in etiolated Arabidopsis cotyledons via interaction with TCP14[J]. Plant Cell, 2019, 31(12): 2996-3014.
doi: 10.1105/tpc.18.00290 URL |
| [38] |
Jin HQ, Xing MG, Cai CM, et al. B-box proteins in Arachis duranensis: genome-wide characterization and expression profiles analysis[J]. Agronomy, 2019, 10(1): 23.
doi: 10.3390/agronomy10010023 URL |
| [39] |
Hartings S, Paradies S, Karnuth B, et al. The DnaJ-like zinc-finger protein HCF222 is required for thylakoid membrane biogenesis in plants[J]. Plant Physiol, 2017, 174(3): 1807-1824.
doi: 10.1104/pp.17.00401 pmid: 28572458 |
| [40] |
Ham BK, Park JM, Lee SB, et al. Tobacco Tsip1, a DnaJ-type Zn finger protein, is recruited to and potentiates Tsi1-mediated transcriptional activation[J]. Plant Cell, 2006, 18(8): 2005-2020.
doi: 10.1105/tpc.106.043158 URL |
| [41] |
Berman J, Zorrilla-López U, Medina V, et al. The Arabidopsis ORANGE(AtOR)gene promotes carotenoid accumulation in transgenic corn hybrids derived from parental lines with limited carotenoid pools[J]. Plant Cell Rep, 2017, 36(6): 933-945.
doi: 10.1007/s00299-017-2126-z pmid: 28314904 |
| [42] |
Yuan H, Owsiany K, Sheeja TE, et al. A single amino acid substitution in an ORANGE protein promotes carotenoid overaccumulation in Arabidopsis[J]. Plant Physiol, 2015, 169(1): 421-431.
doi: 10.1104/pp.15.00971 pmid: 26224804 |
| [43] |
Zhong LL, Zhou W, Wang HJ, et al. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress[J]. Plant Cell, 2013, 25(8): 2925-2943.
doi: 10.1105/tpc.113.111229 URL |
| [44] | Jin HL, Luo LJ, Wang JF, Wang HB. A novel thylakoid protein, HIGH LIGHT SENSITIVITY1, interacts with LQY1 and involves in photoprotection of photosystemⅡ in Arabidopsis thaliana[C]// 2013 National Congress of Plant Biology, Nanjing, 2013: 248. |
| [45] |
Jacob P, Hirt H, Bendahmane A. The heat-shock protein/chaperone network and multiple stress resistance[J]. Plant Biotechnol J, 2017, 15(4): 405-414.
doi: 10.1111/pbi.12659 pmid: 27860233 |
| [46] |
胡慧芳, 王子雨, 潘潇潇, 等. 辣椒转录因子CaNAC083对高温胁迫的响应[J/OL]. 西北农林科技大学学报: 自然科学版, 2023. DOI:10.13207/j.cnki.jnwafu.2023.03.013.
doi: 10.13207/j.cnki.jnwafu.2023.03.013 |
|
Hu HF, Wang ZY, Pan XX, et al. Response of pepper transcription factor CaNAC083 to high temperature stres[J/OL]. J Northwest A & F Univ Nat Sci Ed, 2023. DOI:10.13207/j.cnki.jnwafu.2023.03.013.
doi: 10.13207/j.cnki.jnwafu.2023.03.013 |
|
| [47] |
Sun JT, Cheng GX, Huang LJ, et al. Modified expression of a heat shock protein gene, CaHSP22.0, results in high sensitivity to heat and salt stress in pepper(Capsicum annuum L.)[J]. Sci Hortic, 2019, 249: 364-373.
doi: 10.1016/j.scienta.2019.02.008 URL |
| [48] |
Huang LJ, Cheng GX, Khan A, et al. CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance[J]. Protoplasma, 2019, 256(1): 39-51.
doi: 10.1007/s00709-018-1280-7 URL |
| [49] | 张驰, 刘丹丹, 刘建中. 植物J蛋白的生物学功能及其作用机制[J]. 浙江大学学报: 农业与生命科学版, 2018, 44(3): 275-282. |
| Zhang C, Liu DD, Liu JZ. Biological functions and action mechanisms of J-domain proteins in plants[J]. J Zhejiang Univ Agric Life Sci, 2018, 44(3): 275-282. | |
| [50] |
Lee KW, Rahman MA, Kim KY, et al. Overexpression of the alfalfa DnaJ-like protein(MsDJLP)gene enhances tolerance to chilling and heat stresses in transgenic tobacco plants[J]. Turk J Biol, 2018, 42(1): 12-22.
doi: 10.3906/biy-1705-30 pmid: 30814866 |
| [51] |
So HA, Chung E, Lee JH. Molecular characterization of soybean GmDjp1 encoding a type III J-protein induced by abiotic stress[J]. Genes Genom, 2013, 35(2): 247-256.
doi: 10.1007/s13258-013-0078-4 URL |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | ZHAO Zhi-xiang, WANG Dian-dong, ZHOU Ya-lin, WANG Pei, YAN Wan-rong, YAN Bei, LUO Lu-yun, ZHANG Zhuo. Control of Pepper Fusarium Wilt by Bacillus subtilis Ya-1 and Its Effect on Rhizosphere Fungal Microbial Community [J]. Biotechnology Bulletin, 2023, 39(9): 213-224. |
| [3] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [4] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | YE Yun-fang, TIAN Qing-yin, SHI Ting-ting, WANG Liang, YUE Yuan-zheng, YANG Xiu-lian, WANG Liang-gui. Research Progress in the Biosynthesis and Regulation of β-ionone in Plants [J]. Biotechnology Bulletin, 2023, 39(8): 91-105. |
| [7] | WEI Xi-ya, QIN Zhong-wei, LIANG La-mei, LIN Xin-qi, LI Ying-zhi. Mechanism of Melatonin Seed Priming in Improving Salt Tolerance of Capsicum annuum [J]. Biotechnology Bulletin, 2023, 39(7): 160-172. |
| [8] | ZHANG Bei, REN Fu-sen, ZHAO Yang, GUO Zhi-wei, SUN Qiang, LIU He-juan, ZHEN Jun-qi, WANG Tong-tong, CHENG Xiang-jie. Advances in the Mechanism of Pepper in the Response to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(7): 37-47. |
| [9] | LI Ying, YUE Xiang-hua. Application of DNA Methylation in Interpreting Natural Variation in Moso Bamboo [J]. Biotechnology Bulletin, 2023, 39(7): 48-55. |
| [10] | CHENG Ting, YUAN Shuai, ZHANG Xiao-yuan, LIN Liang-cai, LI Xin, ZHANG Cui-ying. Research Progress in the Regulation of Isobutanol Synthesis Pathway in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(7): 80-90. |
| [11] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [12] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [13] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [14] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [15] | SHI Jian-lei, ZAI Wen-shan, SU Shi-wen, FU Cun-nian, XIONG Zi-li. Identification and Expression Analysis of miRNA Related to Bacterial Wilt Resistance in Tomato [J]. Biotechnology Bulletin, 2023, 39(5): 233-242. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||