








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (2): 63-69.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0848
Previous Articles Next Articles
LU Zhen-wan1,2( ), LI Xue-qi1,2,3, HUANG Jin-guang3, ZHOU Huan-bin1,2(
), LI Xue-qi1,2,3, HUANG Jin-guang3, ZHOU Huan-bin1,2( )
)
Received:2022-07-10
Online:2023-02-26
Published:2023-03-07
LU Zhen-wan, LI Xue-qi, HUANG Jin-guang, ZHOU Huan-bin. Creation of Glyphosate-tolerant Rice by Cytosine Base Editing[J]. Biotechnology Bulletin, 2023, 39(2): 63-69.
| 名称 Name | 抗性/菌株 Resistance/Species | 来源 Source | |
|---|---|---|---|
| Plasmid | pENTR4:sgRNA5 | KanR | Lab stock |
| pUbi:rBE22 | KanR | Lab stock | |
| Strain | JM109 | Escherichia coli | Lab stock |
| EHA105 | Agrobacterium tumefaciens | Lab stock |
Table 1 Bacterial strains and plasmids
| 名称 Name | 抗性/菌株 Resistance/Species | 来源 Source | |
|---|---|---|---|
| Plasmid | pENTR4:sgRNA5 | KanR | Lab stock |
| pUbi:rBE22 | KanR | Lab stock | |
| Strain | JM109 | Escherichia coli | Lab stock |
| EHA105 | Agrobacterium tumefaciens | Lab stock |

Fig. 1 Amino acid sequence alignment of rice OsEPSPS and Eleusine indica EiEPSPS EiEPSPS-R(CAD01095.1)and EiEPSPS-S(CAD01096.1)were isolated from the resistant(R)and sensitive(S)Eleusine indica biotypes, respectively[22]. EiEPSPS-P106 site is highlighted in red box
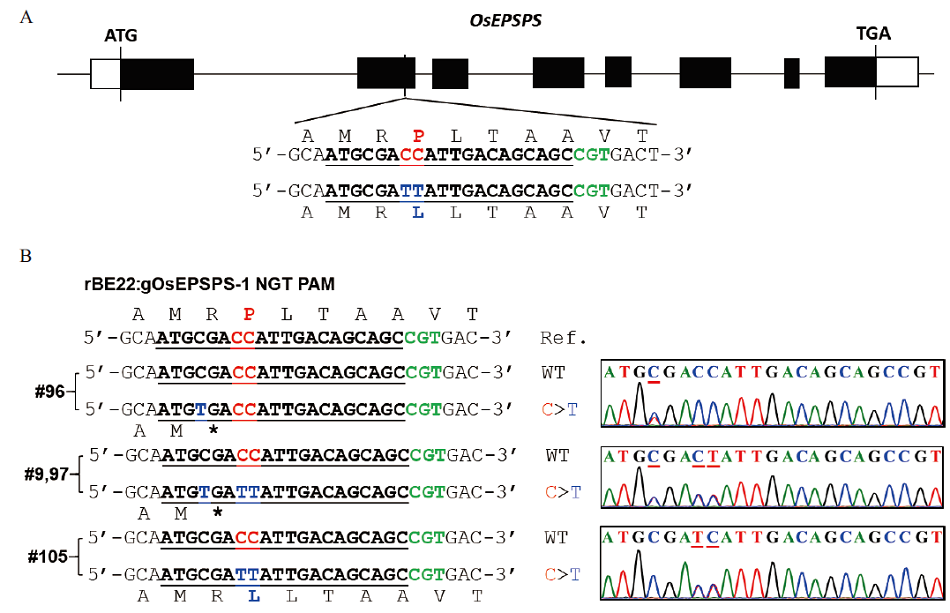
Fig. 2 Identification of base editing materials in T0 A: Design of the rice OsEPSPS gene target site. B: Detected results of the OsEPSPS target base editing in T0 transgenic plants. PAM sequences, target sequences, the target bases and target amino acids, and the mutated predicted bases and corresponding amino acids are highlighted in green, bold, red, and blue, respectively. The mutated bases in the sequencing chromatograms are underlined
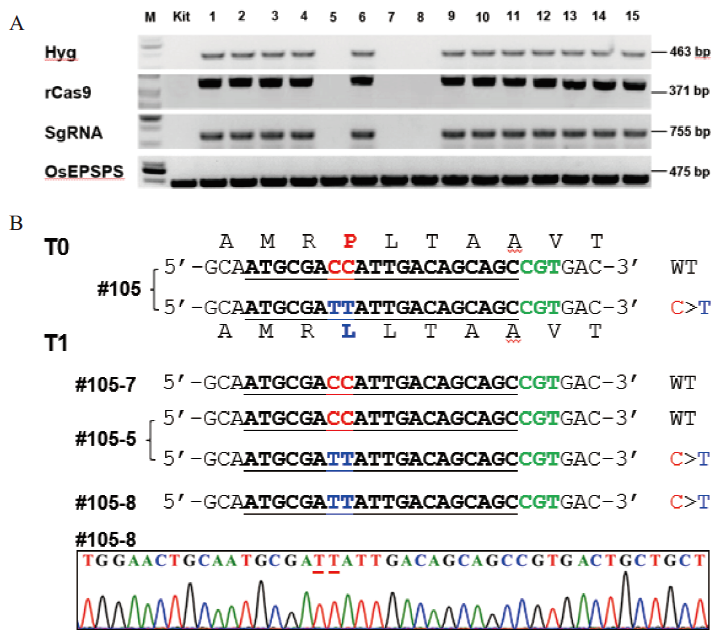
Fig. 3 Identification of base editing materials OsEPSPS(P177L)without exogenous components and homozygous mutations A: Separation detection of exogenous T-DNA elements(sgRNA, Hyg and Cas9)from self-progeny of base editing material #105. Kit is wild-type Kitaake, and lane 1-15 is the T1 plants of #105. B: Sequencing results of the target sites in transgene-free T1 progeny plants. PAM sequences, target sequence, the target bases and target amino acids, and the mutated predicted bases/corresponding amino acids are highlighted in green, bold, red, and blue, respectively. The mutated bases in the sequencing chromatograms are underlined

Fig. 4 Glyphosate-tolerance assay of base editing materials OsEPSPS(P177L)plants A: Germination assay of OsEPSPS(P177L)seeds in cylinders containing 1/2MS with glyphosate, and photos were taken after 14 d treatment. Bar=5 cm. B: Glyphosate-tolerance assay of OsEPSPS(P177L)seedling by spraying glyphosate with spray tower, photographs were taken at 14 d respectively,the recommended dose of glyphosate in the field was 1 350 g a.i./ha as 1×
| [1] |
Nguyen N, Ferrero A. Meeting the challenges of global rice production[J]. Paddy Water Environ, 2006, 4(1): 1-9.
doi: 10.1007/s10333-005-0031-5 URL |
| [2] |
Farooq M, Siddique KHM, Rehman H, et al. Rice direct seeding: Experiences, challenges and opportunities[J]. Soil Tillage Res, 2011, 111(2): 87-98.
doi: 10.1016/j.still.2010.10.008 URL |
| [3] | Akbar N, Ehsanullah, Jabran K, et al. Weed management improves yield and quality of direct seeded rice[J]. Australian Journal of Crop Science, 2011, 5(6): 688-694. |
| [4] |
Singh Y, Singh VP, Singh G, et al. The implications of land preparation, crop establishment method and weed management on rice yield variation in the rice-wheat system in the Indo-Gangetic plains[J]. Field Crops Res, 2011, 121(1): 64-74.
doi: 10.1016/j.fcr.2010.11.012 URL |
| [5] |
Chauhan BS. Effect of tillage systems, seeding rates, and herbicides on weed growth and grain yield in dry-seeded rice systems in the Philippines[J]. Crop Prot, 2013, 54: 244-250.
doi: 10.1016/j.cropro.2013.09.001 URL |
| [6] |
Antralina M, Istina IN, Yuwariah Y, et al. Effect of difference weed control methods to yield of lowland rice in the SOBARI[J]. Procedia Food Sci, 2015, 3: 323-329.
doi: 10.1016/j.profoo.2015.01.035 URL |
| [7] |
Chauhan BS, Awan TH, Abugho SB, et al. Effect of crop establishment methods and weed control treatments on weed management, and rice yield[J]. Field Crops Res, 2015, 172: 72-84.
doi: 10.1016/j.fcr.2014.12.011 URL |
| [8] |
Duke SO, Powles SB. Glyphosate: a once-in-a-century herbicide[J]. Pest Manag Sci, 2008, 64(4): 319-325.
doi: 10.1002/ps.1518 pmid: 18273882 |
| [9] |
Steinrücken HC, Amrhein N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase[J]. Biochem Biophys Res Commun, 1980, 94(4): 1207-1212.
doi: 10.1016/0006-291X(80)90547-1 URL |
| [10] |
Herrmann KM. The shikimate pathway: early steps in the biosynthesis of aromatic compounds[J]. Plant Cell, 1995, 7(7): 907-919.
doi: 10.2307/3870046 URL |
| [11] |
Roberts F, Roberts CW, Johnson JJ, et al. Evidence for the shikimate pathway in apicomplexan parasites[J]. Nature, 1998, 393(6687): 801-805.
doi: 10.1038/31723 URL |
| [12] |
Richards TA, Dacks JB, Campbell SA, et al. Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements[J]. Eukaryot Cell, 2006, 5(9): 1517-1531.
pmid: 16963634 |
| [13] |
Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424.
doi: 10.1038/nature17946 URL |
| [14] |
Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471.
doi: 10.1038/nature24644 URL |
| [15] |
Ren B, Yan F, Kuang YJ, et al. A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice[J]. Sci China Life Sci, 2017, 60(5): 516-519.
doi: 10.1007/s11427-016-0406-x pmid: 28260228 |
| [16] |
Yan F, Kuang YJ, Ren B, et al. Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice[J]. Mol Plant, 2018, 11(4): 631-634.
doi: S1674-2052(18)30058-3 pmid: 29476918 |
| [17] |
Kuang YJ, Li SF, Ren B, et al. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms[J]. Mol Plant, 2020, 13(4): 565-572.
doi: S1674-2052(20)30010-1 pmid: 32001363 |
| [18] |
Zhang R, Liu JX, Chai ZZ, et al. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing[J]. Nat Plants, 2019, 5(5): 480-485.
doi: 10.1038/s41477-019-0405-0 pmid: 30988404 |
| [19] |
Tian SW, Jiang LJ, Cui XX, et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing[J]. Plant Cell Rep, 2018, 37(9): 1353-1356.
doi: 10.1007/s00299-018-2299-0 pmid: 29797048 |
| [20] |
Liu XS, Qin RY, Li J, et al. A CRISPR-Cas9-mediated domain-specific base-editing screen enables functional assessment of ACCase variants in rice[J]. Plant Biotechnol J, 2020, 18(9): 1845-1847.
doi: 10.1111/pbi.13348 pmid: 31985873 |
| [21] |
Liu L, Kuang YJ, Yan F, et al. Developing a novel artificial rice germplasm for dinitroaniline herbicide resistance by base editing of OsTubA2[J]. Plant Biotechnol J, 2021, 19(1): 5-7.
doi: 10.1111/pbi.13430 URL |
| [22] |
Baerson SR, Rodriguez DJ, Tran M, et al. Glyphosate-resistant goosegrass. Identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase[J]. Plant Physiol, 2002, 129(3): 1265-1275.
pmid: 12114580 |
| [23] |
Zhou HB, Liu B, Weeks DP, et al. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice[J]. Nucleic Acids Res, 2014, 42(17): 10903-10914.
doi: 10.1093/nar/gku806 pmid: 25200087 |
| [24] |
Hiei Y, Ohta S, Komari T, et al. Efficient transformation of rice(Oryza sativa L.)mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA[J]. Plant J, 1994, 6(2): 271-282.
doi: 10.1046/j.1365-313x.1994.6020271.x pmid: 7920717 |
| [25] | Cui Y, Huang SQ, Liu ZD, et al. Development of novel glyphosate-tolerant Japonica rice lines: a step toward commercial release[J]. Front Plant Sci, 2016, 7: 1218. |
| [1] | LI Xue-qi, ZHANG Su-jie, YU Man, HUANG Jin-guang, ZHOU Huan-bin. Establishment of CRISPR/CasX-based Genome Editing Technology in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 40-48. |
| [2] | LIU Jia-hui, LIU Ye, HUA Er-bing, WANG Meng. PAM Extension of Cytosine Base Editing Tool in Corynebacterium glutamicum [J]. Biotechnology Bulletin, 2023, 39(9): 49-57. |
| [3] | LI Peng-cheng, ZHANG Ming-jun, WANG Yin-xiao, LI Xiang-yin, LI Sheng-yan, LANG Zhi-hong. Insect Resistance Identification and Agronomy Traits Analysis of Transgenic Maize HGK60 with Different Genetic Backgrounds [J]. Biotechnology Bulletin, 2023, 39(1): 40-47. |
| [4] | WANG Cui-cui, FU Da-qi. Research Progress in the Effects of Ubiquitin-proteasome System on Plant Agronomic Traits [J]. Biotechnology Bulletin, 2023, 39(1): 72-83. |
| [5] | LIANG Hai-sheng, LI Meng-tao, LI Sheng-yan, WANG Hai, ZHANG Jie, LANG Zhi-hong. Agronomic Traits Analysis of Transgenic Bt cry1Ah Maize HGK60 Line [J]. Biotechnology Bulletin, 2018, 34(7): 92-100. |
| [6] | SONG Yan-chao, An Fei-fei, Xue Jing-jing, Qin Yu-ling, Li Kai-mian, CHEN Song-bi. Proteomic Analysis on Tuberous Roots of Cassava Cultivar ZM-Seaside and Mosaic-leaf Mutation [J]. Biotechnology Bulletin, 2017, 33(3): 78-85. |
| [7] | YANG Hui-zhen, REN Zhi-qiang, XIAO Jian-hong, BU Hua-hu, LIU Hui-min. Development of Transgenic Maize Plants Tolerant to Glyphosate via Pollen-mediated Transformation and Their Glyphosate Tolerance [J]. Biotechnology Bulletin, 2016, 32(11): 152-161. |
| [8] | Dai Jun, Li Xiuying, Zhu Li, Wang Hai, Zhang Jie, He Kanglai, Lang Zhihong, Huang Dafang. Molecular Detection and Agronomic Traits Analysis of Insect-resistant Transgenic Maize Harboring Bt cry1Ah Gene [J]. Biotechnology Bulletin, 2014, 0(5): 62-68. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||