








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (3): 35-42.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0816
Previous Articles Next Articles
CUI Jun-mei1( ), WEI Jia-ping1, DONG Xiao-yun2, WANG Ying2, ZHENG Guo-qiang2, LIU Zi-gang1(
), WEI Jia-ping1, DONG Xiao-yun2, WANG Ying2, ZHENG Guo-qiang2, LIU Zi-gang1( )
)
Received:2022-07-01
Online:2023-03-26
Published:2023-04-10
CUI Jun-mei, WEI Jia-ping, DONG Xiao-yun, WANG Ying, ZHENG Guo-qiang, LIU Zi-gang. PIP/PIPL: A Kind of Endogenous Plant Peptide Regulating Plant Stress Response and Development[J]. Biotechnology Bulletin, 2023, 39(3): 35-42.
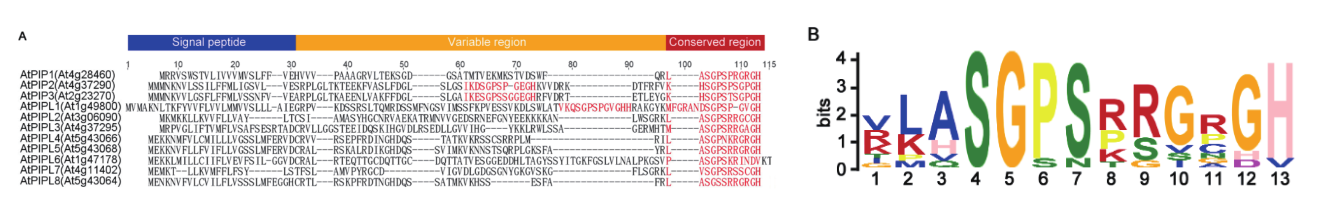
Fig. 1 Sequence alignment and conserved motif of AtPIP/PIPL family proteins A: Alignment based on full-length sequence of AtPIP/PIPL precursor proteins. The conserved motifs were marked in red. B: Conserved sequence features of AtPIP/PIPL
| [1] |
Matsubayashi Y, Sakagami Y. Peptide hormones in plants[J]. Annu Rev Plant Biol, 2006, 57: 649-674.
pmid: 16669777 |
| [2] |
Matsubayashi Y. Posttranslationally modified small-peptide signals in plants[J]. Annu Rev Plant Biol, 2014, 65: 385-413.
doi: 10.1146/annurev-arplant-050312-120122 pmid: 24779997 |
| [3] |
Lease KA, Walker JC. Bioinformatic identification of plant peptides[J]. Methods Mol Biol, 2010, 615: 375-383.
doi: 10.1007/978-1-60761-535-4_26 pmid: 20013221 |
| [4] |
Lease KA, Walker JC. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics[J]. Plant Physiol, 2006, 142(3): 831-838.
doi: 10.1104/pp.106.086041 URL |
| [5] |
Takahashi F, Suzuki T, Osakabe Y, et al. A small peptide modulates stomatal control via abscisic acid in long-distance signalling[J]. Nature, 2018, 556(7700): 235-238.
doi: 10.1038/s41586-018-0009-2 |
| [6] |
Takahashi F, Hanada K, Kondo T, et al. Hormone-like peptides and small coding genes in plant stress signaling and development[J]. Curr Opin Plant Biol, 2019, 51: 88-95.
doi: S1369-5266(19)30028-7 pmid: 31265991 |
| [7] |
Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response[J]. PNAS, 2006, 103(26): 10098-10103.
doi: 10.1073/pnas.0603727103 pmid: 16785434 |
| [8] |
Stegmann M, Monaghan J, Smakowska-Luzan E, et al. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling[J]. Science, 2017, 355(6322): 287-289.
doi: 10.1126/science.aal2541 pmid: 28104890 |
| [9] |
Zhou HP, Xiao F, Zheng Y, et al. Pamp-induced secreted peptide 3 modulates salt tolerance through receptor-like kinase 7 in plants[J]. Plant Cell, 2022, 34(2): 927-944.
doi: 10.1093/plcell/koab292 URL |
| [10] |
Najafi J, Brembu T, Vie AK, et al. Pamp-induced secreted peptide 3(pip3)modulates immunity in Arabidopsis thaliana[J]. J Exp Bot, 2020, 71(3):850-864.
doi: 10.1093/jxb/erz482 pmid: 31665431 |
| [11] |
Hussain S, Wang W, Ahmed S, et al. PIP2, an auxin induced plant peptide hormone regulates root and hypocotyl elongation in Arabidopsis[J]. Front Plant Sci, 2021, 12: 646736.
doi: 10.3389/fpls.2021.646736 URL |
| [12] |
Hou SG, Wang X, Chen DH, et al. The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7[J]. PLoS Pathog, 2014, 10(9): e1004331.
doi: 10.1371/journal.ppat.1004331 URL |
| [13] |
Vie AK, Najafi J, Liu B, et al. The IDA/IDA-LIKE and PIP/PIP-LIKE gene families in Arabidopsis: phylogenetic relationship, expression patterns, and transcriptional effect of the PIPL3 peptide[J]. J Exp Bot, 2015, 66(17): 5351-5365.
doi: 10.1093/jxb/erv285 URL |
| [14] |
Combest MM, Moroz N, Tanaka K, et al. StPIP1, a PAMP-induced peptide in potato, elicits plant defenses and is associated with disease symptom severity in a compatible interaction with Potato virus Y[J]. J Exp Bot, 2021, 72(12): 4472-4488.
doi: 10.1093/jxb/erab078 pmid: 33681961 |
| [15] |
Ito Y, Nakanomyo I, Motose H, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation[J]. Science, 2006, 313(5788): 842-845.
doi: 10.1126/science.1128436 pmid: 16902140 |
| [16] |
Meng L, Buchanan BB, Feldman LJ, et al. CLE-like(CLEL)peptides control the pattern of root growth and lateral root development in Arabidopsis[J]. Proc Natl Acad Sci USA, 2012, 109(5): 1760-1765.
doi: 10.1073/pnas.1119864109 URL |
| [17] |
Aalen RB, Wildhagen M, Stø IM, et al. IDA: a peptide ligand regulating cell separation processes in Arabidopsis[J]. J Exp Bot, 2013, 64(17): 5253-5261.
doi: 10.1093/jxb/ert338 URL |
| [18] |
Zipfel C. Plant pattern-recognition receptors[J]. Trends Immunol, 2014, 35(7): 345-351.
doi: 10.1016/j.it.2014.05.004 pmid: 24946686 |
| [19] |
Belkhadir Y, Yang L, Hetzel J, et al. The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors[J]. Trends Biochem Sci, 2014, 39(10): 447-456.
doi: 10.1016/j.tibs.2014.06.006 pmid: 25089011 |
| [20] |
Jones JDG, Dangl JL. The plant immune system[J]. Nature, 2006, 444(7117): 323-329.
doi: 10.1038/nature05286 |
| [21] |
Gómez-Gómez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis[J]. Mol Cell, 2000, 5(6): 1003-1011.
doi: 10.1016/s1097-2765(00)80265-8 pmid: 10911994 |
| [22] |
Böhm H, et al. A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis[J]. PLoS Pathog, 2014, 10(11): e1004491.
doi: 10.1371/journal.ppat.1004491 URL |
| [23] |
Sakamoto T, Deguchi M, Brustolini OJB, et al. The tomato RLK superfamily: phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense[J]. BMC Plant Biol, 2012, 12: 229.
doi: 10.1186/1471-2229-12-229 pmid: 23198823 |
| [24] |
Dunning FM, Sun WX, Jansen KL, et al. Identification and mutational analysis of Arabidopsis FLS2 leucine-rich repeat domain residues that contribute to flagellin perception[J]. Plant Cell, 2007, 19(10): 3297-3313.
doi: 10.1105/tpc.106.048801 pmid: 17933906 |
| [25] |
Tang J, Han ZF, Sun YD, et al. Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1[J]. Cell Res, 2015, 25(1): 110-120.
doi: 10.1038/cr.2014.161 pmid: 25475059 |
| [26] |
Zhang XX, Liu WJ, Nagae TT, et al. Structural basis for receptor recognition of pollen tube attraction peptides[J]. Nat Commun, 2017, 8(1): 1331.
doi: 10.1038/s41467-017-01323-8 pmid: 29109411 |
| [27] |
Hallgren J, Tsirigos KD, Pedersen MD, et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks[J]. bioRxiv, 2022, DOI:10.1101/2022.04.08.487609.
doi: 10.1101/2022.04.08.487609 |
| [28] |
Chen TS. Identification and characterization of the LRR repeats in plant LRR-RLKs[J]. BMC Mol Cell Biol, 2021, 22(1): 9.
doi: 10.1186/s12860-021-00344-y pmid: 33509084 |
| [29] |
Liebrand TWH, van den Burg HA, Joosten MHAJ. Two for all: receptor-associated kinases SOBIR1 and BAK1[J]. Trends Plant Sci, 2014, 19(2): 123-132.
doi: 10.1016/j.tplants.2013.10.003 pmid: 24238702 |
| [30] |
Roux M, Schwessinger B, Albrecht C, et al. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens[J]. Plant Cell, 2011, 23(6): 2440-2455.
doi: 10.1105/tpc.111.084301 URL |
| [31] |
Liang XX, Zhou JM. Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling[J]. Annu Rev Plant Biol, 2018, 69: 267-299.
doi: 10.1146/annurev-arplant-042817-040540 pmid: 29719165 |
| [32] |
Zhang J, Li W, Xiang TT, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector[J]. Cell Host Microbe, 2010, 7(4): 290-301.
doi: 10.1016/j.chom.2010.03.007 pmid: 20413097 |
| [33] |
Lei JX, Finlayson SA, Salzman RA, et al. BOTRYTIS-INDUCED KINASE1 modulates Arabidopsis resistance to green peach aphids via PHYTOALEXIN DEFICIENT4[J]. Plant Physiol, 2014, 165(4): 1657-1670.
doi: 10.1104/pp.114.242206 URL |
| [34] |
Lu DP, Wu SJ, Gao XQ, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity[J]. PNAS, 2010, 107(1): 496-501.
doi: 10.1073/pnas.0909705107 pmid: 20018686 |
| [35] |
Liu Z, Wu Y, Yang F, et al. BIK1 interacts with PEPRs to mediate ethylene-induced immunity[J]. Proc Natl Acad Sci USA, 2013, 110(15): 6205-6210.
doi: 10.1073/pnas.1215543110 pmid: 23431184 |
| [36] |
Macho AP, Lozano-Durán R, Zipfel C. Importance of tyrosine phosphorylation in receptor kinase complexes[J]. Trends Plant Sci, 2015, 20(5): 269-272.
doi: S1360-1385(15)00050-3 pmid: 25795237 |
| [37] |
Macho AP, Schwessinger B, Ntoukakis V, et al. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation[J]. Science, 2014, 343(6178): 1509-1512.
doi: 10.1126/science.1248849 pmid: 24625928 |
| [38] |
Perraki A, DeFalco TA, Derbyshire P, et al. Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling[J]. Nature, 2018, 561(7722): 248-252.
doi: 10.1038/s41586-018-0471-x |
| [39] |
Chinchilla D, Zipfel C, Robatzek S, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence[J]. Nature, 2007, 448(7152): 497-500.
doi: 10.1038/nature05999 |
| [40] |
Hartmann J, Fischer C, et al. Kinase activity and calmodulin binding are essential for growth signaling by the phytosulfokine receptor PSKR1[J]. Plant J, 2014, 78(2): 192-202.
doi: 10.1111/tpj.12460 URL |
| [41] |
Hou SG, Shen HX, Shao HW. PAMP-induced peptide 1 cooperates with salicylic acid to regulate stomatal immunity in Arabidopsis thaliana[J]. Plant Signal Behav, 2019, 14(11): 1666657.
doi: 10.1080/15592324.2019.1666657 URL |
| [42] |
Yamaguchi Y, Pearce G, Ryan CA. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells[J]. Proc Natl Acad Sci USA, 2006, 103(26): 10104-10109.
pmid: 16785433 |
| [43] |
Yamaguchi Y, Huffaker A, et al. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis[J]. Plant Cell, 2010, 22(2): 508-522.
doi: 10.1105/tpc.109.068874 URL |
| [44] |
Park CH, Bi Y, et al. Deconvoluting signals downstream of growth and immune receptor kinases by phosphocodes of the BSU1 family phosphatases[J]. Nat Plants, 2022, 8(6): 646-655.
doi: 10.1038/s41477-022-01167-1 pmid: 35697730 |
| [45] |
Toyokura K, Goh T, Shinohara H, et al. Lateral inhibition by a peptide hormone-receptor cascade during Arabidopsis lateral root founder cell formation[J]. Dev Cell, 2019, 48(1): 64-75.e5.
doi: S1534-5807(18)30987-0 pmid: 30581155 |
| [46] |
Okushima Y, Fukaki H, Onoda M, et al. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis[J]. Plant Cell, 2007, 19(1): 118-130.
doi: 10.1105/tpc.106.047761 pmid: 17259263 |
| [47] |
Jing YP, Zheng XJ, et al. Danger-associated peptides interact with PIN-dependent local auxin distribution to inhibit root growth in Arabidopsis[J]. Plant Cell, 2019, 31(8): 1767-1787.
doi: 10.1105/tpc.18.00757 URL |
| [1] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [2] | LIU Wen-jin, MA Rui, LIU Sheng-yan, YANG Jiang-wei, ZHANG Ning, SI Huai-jun. Cloning of StCIPK11 Gene and Analysis of Its Response to Drought Stress in Solanum tuberosum [J]. Biotechnology Bulletin, 2023, 39(9): 147-155. |
| [3] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [4] | KANG Ling-yun, HAN Lu-lu, HAN De-ping, CHEN Jian-sheng, GAN Han-ling, XING Kai, MA You-ji, CUI Kai. Effect of Melatonin on Protecting the Jejunum Mucosal Epithelial Cells from Oxidative Stress Damage [J]. Biotechnology Bulletin, 2023, 39(9): 291-299. |
| [5] | JIANG Run-hai, JIANG Ran-ran, ZHU Cheng-qiang, HOU Xiu-li. Research Progress in Mechanisms of Microbial-enhanced Phytoremediation for Lead-contaminated Soil [J]. Biotechnology Bulletin, 2023, 39(8): 114-125. |
| [6] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [7] | HU Hai-lin, XU Li, LI Xiao-xu, WANG Chen-can, MEI Man, DING Wen-jing, ZHAO Yuan-yuan. Advances in the Regulation of Plant Growth, Development and Stress Physiology by Small Peptide Hormones [J]. Biotechnology Bulletin, 2023, 39(7): 13-25. |
| [8] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [9] | WEI Xi-ya, QIN Zhong-wei, LIANG La-mei, LIN Xin-qi, LI Ying-zhi. Mechanism of Melatonin Seed Priming in Improving Salt Tolerance of Capsicum annuum [J]. Biotechnology Bulletin, 2023, 39(7): 160-172. |
| [10] | YU Hui, WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun. Isolation and Functional Verification of Genes Responding to Iron and Cadmium Stresses in Lycium barbarum [J]. Biotechnology Bulletin, 2023, 39(7): 195-205. |
| [11] | ZHANG Bei, REN Fu-sen, ZHAO Yang, GUO Zhi-wei, SUN Qiang, LIU He-juan, ZHEN Jun-qi, WANG Tong-tong, CHENG Xiang-jie. Advances in the Mechanism of Pepper in the Response to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(7): 37-47. |
| [12] | LI Ying, YUE Xiang-hua. Application of DNA Methylation in Interpreting Natural Variation in Moso Bamboo [J]. Biotechnology Bulletin, 2023, 39(7): 48-55. |
| [13] | DING Kai-xin, WANG Li-chun, TIAN Guo-kui, WANG Hai-yan, LI Feng-yun, PAN Yang, PANG Ze, SHAN Ying. Research Progress in Uniconazole Alleviating Plant Drought Damage [J]. Biotechnology Bulletin, 2023, 39(6): 1-11. |
| [14] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [15] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||