








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (6): 171-180.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1341
Previous Articles Next Articles
MENG Guo-qiang( ), GUAN Jian-wen, NIU Chun-mei, ZHOU Ying, SHEN Su-lin, WEI You-heng(
), GUAN Jian-wen, NIU Chun-mei, ZHOU Ying, SHEN Su-lin, WEI You-heng( )
)
Received:2022-11-01
Online:2023-06-26
Published:2023-07-07
Contact:
WEI You-heng
E-mail:m17854338552@163.com;yhwei@yzu.edu.cn
MENG Guo-qiang, GUAN Jian-wen, NIU Chun-mei, ZHOU Ying, SHEN Su-lin, WEI You-heng. Construction and Functional Study of RagA Transgenic Drosophila[J]. Biotechnology Bulletin, 2023, 39(6): 171-180.
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| RagA-F | AGTCACTATGGCGGCCGCCATGAAGAAAAAGGTGTTAC |
| RagA-R | GATGCGGCCTCCACCGCGGCAATGGTACCTTTGGCCATG |
| M-F | ATCAGAAGCAACACTTGTAAACATAAGGAACGCTC |
| M-R | TTTACAAGTGTTGCTTCTGATGGCAGCGTGGGATCCG |
| Q61L-F | ACTGTGGCGGTCTGGAGGGCTTC |
| Q61L-R | GAAGCCCTCCAGACCGCCACAGT |
| T16N-F | CCGGAAAGAACAGCATGCGCTC |
| T16N-R | GAGCGCATGCTGTTCTTTCCGG |
| RagA-qPCR-F | GCCAGAGCAAGAAGAACC |
| RagA-qPCR-R | CAATGAAAGCGGCAAAT |
| rp49-qPCR-F | GCCGCTTCAAGGGACAGT |
| rp49-qPCR-R | CGATCTCGCCGCAGTAAA |
Table 1 PCR primer sequences
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| RagA-F | AGTCACTATGGCGGCCGCCATGAAGAAAAAGGTGTTAC |
| RagA-R | GATGCGGCCTCCACCGCGGCAATGGTACCTTTGGCCATG |
| M-F | ATCAGAAGCAACACTTGTAAACATAAGGAACGCTC |
| M-R | TTTACAAGTGTTGCTTCTGATGGCAGCGTGGGATCCG |
| Q61L-F | ACTGTGGCGGTCTGGAGGGCTTC |
| Q61L-R | GAAGCCCTCCAGACCGCCACAGT |
| T16N-F | CCGGAAAGAACAGCATGCGCTC |
| T16N-R | GAGCGCATGCTGTTCTTTCCGG |
| RagA-qPCR-F | GCCAGAGCAAGAAGAACC |
| RagA-qPCR-R | CAATGAAAGCGGCAAAT |
| rp49-qPCR-F | GCCGCTTCAAGGGACAGT |
| rp49-qPCR-R | CGATCTCGCCGCAGTAAA |
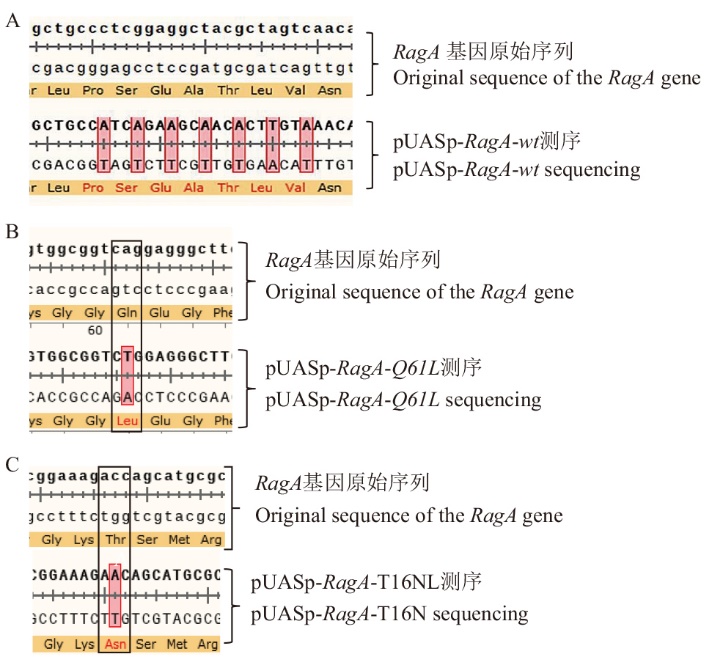
Fig. 1 Construction of recombinant plasmid A: DNA sequencing comparison chart of pUASp-RagA-wt recombinant plasmid; B: DNA sequencing comparison chart of pUASp-RagA-Q61L recombinant plasmid; C: DNA sequencing comparison chart of pUASp-RagA-T16N recombinant plasmid
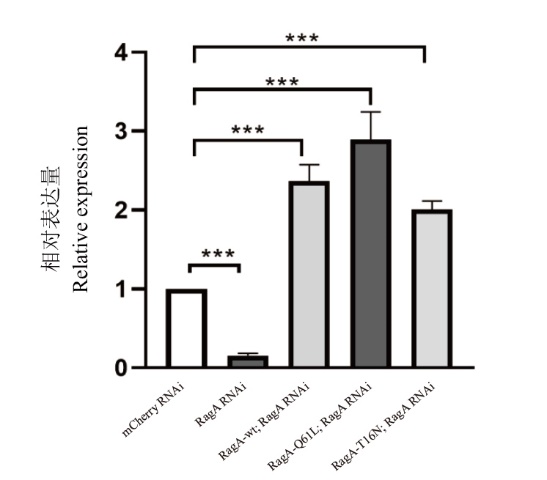
Fig. 3 qPCR detection of RagA gene expression mCherry RNAi is the control group; RagA RNAi is RagA RNA interference; RagA-wt; RagA RNAi is an overexpression of wild-type RagA based on RagA RNA interference; RagA-Q61L; RagA RNAi is RagA overexpressing GTP-bound state on the basis of RagA RNA interference; RagA-T16N; RagA RNAi is RagA overexpressing GDP-bound state on the basis of RagA RNA interference; ***: P<0.000 5; ****: P<0.000 1. The same below
| 基因型 Genotype | 羽化数 Eclosion number | 实际比例 Actual prop- ortions/% | 理论比例 Theoretical proportions/% |
|---|---|---|---|
| TM6/UAS-RagA RNAi | 897 | 100 | 50 |
| Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 50 |
| 总数 Total | 897 | 100 | 100 |
Table 2 RagA knockdown on the statistical results of Drosophila eclosion rate
| 基因型 Genotype | 羽化数 Eclosion number | 实际比例 Actual prop- ortions/% | 理论比例 Theoretical proportions/% |
|---|---|---|---|
| TM6/UAS-RagA RNAi | 897 | 100 | 50 |
| Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 50 |
| 总数 Total | 897 | 100 | 100 |

Fig. 4 Effects of RagA knockdown on Drosophila at different developmental stages A: Ratios from larval to pupal stages in control and RagA knockdown drosophilas. B: Eclosion rate from pupal stage to adult in control and RagA knockdown drosophilas. Error bars represent the standard deviation of 10 independent experiments. C: Developmental map of RagA knockdown and control flies. n refers to the total number of statistics. NS: No significant difference

Fig. 5 Somatic cell cloning assay for cell size A-A’’: RagA RNAi somatic cell clone; B-B’’: RagA-Q61L; RagA RNAi somatic cell clone; C-C’’: RagA-T16N; RagA RNAi somatic cell clone; D: RagA RNAi clone cell size statistics; E: RagA-Q61L; RagA RNAi clone cell size statistics; F: RagA-T16N; RagA RNAi clone cell size statistics
| 基因型 Genotype | 羽化数 Eclosion number | 实际比例 Actual pro- portion/% | 理论比例 Theoretical proportion/% |
|---|---|---|---|
| UAS-RagA-wt /+; Tub-Gal4/UAS-RagA RNAi | 214 | 14.8 | 12.5 |
| UAS-RagA-wt /+; Tub-Gal4/MKRS | 220 | 15.3 | 12.5 |
| UAS-RagA-wt /+; MKRS/TM6 | 204 | 14.1 | 12.5 |
| UAS-RagA-wt /+; UAS-RagA RNAi/TM6 | 199 | 13.8 | 12.5 |
| SM6/+; Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 12.5 |
| SM6/+; Tub-Gal4/MKRS | 192 | 13.4 | 12.5 |
| SM6/+; MKRS/TM6 | 212 | 14.7 | 12.5 |
| SM6/+; UAS-RagA RNAi/TM6 | 201 | 13.9 | 12.5 |
| 总数 Total | 1 442 | 100 | 100 |
Table 3 Statistical results of the lethal effect of RagA overexpression on Drosophila
| 基因型 Genotype | 羽化数 Eclosion number | 实际比例 Actual pro- portion/% | 理论比例 Theoretical proportion/% |
|---|---|---|---|
| UAS-RagA-wt /+; Tub-Gal4/UAS-RagA RNAi | 214 | 14.8 | 12.5 |
| UAS-RagA-wt /+; Tub-Gal4/MKRS | 220 | 15.3 | 12.5 |
| UAS-RagA-wt /+; MKRS/TM6 | 204 | 14.1 | 12.5 |
| UAS-RagA-wt /+; UAS-RagA RNAi/TM6 | 199 | 13.8 | 12.5 |
| SM6/+; Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 12.5 |
| SM6/+; Tub-Gal4/MKRS | 192 | 13.4 | 12.5 |
| SM6/+; MKRS/TM6 | 212 | 14.7 | 12.5 |
| SM6/+; UAS-RagA RNAi/TM6 | 201 | 13.9 | 12.5 |
| 总数 Total | 1 442 | 100 | 100 |
| 基因型 Genotype | 羽化数 Eclosion number | 实际比例 Actual pro- portions/% | 理论比例 Theoretical proportions/% |
|---|---|---|---|
| UAS-RagA-Q61L/+; Tub-Gal4/UAS-RagA RNAi | 63 | 4.2 | 12.5 |
| UAS-RagA-Q61L/+; Tub-Gal4/MKRS | 251 | 16.6 | 12.5 |
| UAS-RagA-Q61L/+; MKRS/TM6 | 253 | 16.7 | 12.5 |
| UAS-RagA-Q61L/+; UAS-RagA RNAi/TM6 | 230 | 15.2 | 12.5 |
| SM6/+; Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 12.5 |
| SM6/+; Tub-Gal4/MKRS | 247 | 16.3 | 12.5 |
| SM6/+; MKRS/TM6 | 224 | 14.9 | 12.5 |
| SM6/+; UAS-RagA RNAi/TM6 | 244 | 16.1 | 12.5 |
| 总数 Total | 1 512 | 100 | 100 |
Table 4 Statistical results of the lethal effect of RagA and GTP binding on Drosophila
| 基因型 Genotype | 羽化数 Eclosion number | 实际比例 Actual pro- portions/% | 理论比例 Theoretical proportions/% |
|---|---|---|---|
| UAS-RagA-Q61L/+; Tub-Gal4/UAS-RagA RNAi | 63 | 4.2 | 12.5 |
| UAS-RagA-Q61L/+; Tub-Gal4/MKRS | 251 | 16.6 | 12.5 |
| UAS-RagA-Q61L/+; MKRS/TM6 | 253 | 16.7 | 12.5 |
| UAS-RagA-Q61L/+; UAS-RagA RNAi/TM6 | 230 | 15.2 | 12.5 |
| SM6/+; Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 12.5 |
| SM6/+; Tub-Gal4/MKRS | 247 | 16.3 | 12.5 |
| SM6/+; MKRS/TM6 | 224 | 14.9 | 12.5 |
| SM6/+; UAS-RagA RNAi/TM6 | 244 | 16.1 | 12.5 |
| 总数 Total | 1 512 | 100 | 100 |
| 基因型 Genotype | 羽化数 Eclosion number | 实际比例 Actual pro- portion/% | 理论比例 Theoretical proportion/% |
|---|---|---|---|
| UAS-RagA-T16N/+; Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 12.5 |
| UAS-RagA-T16N/+; Tub-Gal4/MKRS | 240 | 17.2 | 12.5 |
| UAS-RagA-T16N/+; MKRS/TM6 | 228 | 16.3 | 12.5 |
| UAS-RagA-T16N/+; UAS-RagA RNAi/TM6 | 217 | 15.5 | 12.5 |
| SM6/+; Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 12.5 |
| SM6/+; Tub-Gal4/MKRS | 248 | 17.8 | 12.5 |
| SM6/+; MKRS/TM6 | 235 | 16.8 | 12.5 |
| SM6/+; UAS-RagA RNAi/TM6 | 229 | 16.4 | 12.5 |
| 总数 Total | 1 397 | 100 | 100 |
Table 5 Statistical results of the lethal effect of RagA and GDP binding on Drosophila
| 基因型 Genotype | 羽化数 Eclosion number | 实际比例 Actual pro- portion/% | 理论比例 Theoretical proportion/% |
|---|---|---|---|
| UAS-RagA-T16N/+; Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 12.5 |
| UAS-RagA-T16N/+; Tub-Gal4/MKRS | 240 | 17.2 | 12.5 |
| UAS-RagA-T16N/+; MKRS/TM6 | 228 | 16.3 | 12.5 |
| UAS-RagA-T16N/+; UAS-RagA RNAi/TM6 | 217 | 15.5 | 12.5 |
| SM6/+; Tub-Gal4/UAS-RagA RNAi | 0 | 0 | 12.5 |
| SM6/+; Tub-Gal4/MKRS | 248 | 17.8 | 12.5 |
| SM6/+; MKRS/TM6 | 235 | 16.8 | 12.5 |
| SM6/+; UAS-RagA RNAi/TM6 | 229 | 16.4 | 12.5 |
| 总数 Total | 1 397 | 100 | 100 |
| [1] | Frasch M. Genome-wide approaches to Drosophila heart development[J]. J Cardiovasc Dev Dis, 2016, 3(2): 20. |
| [2] |
Adams MD, Celniker SE, Holt RA, et al. The genome sequence of Drosophila melanogaster[J]. Science, 2000, 287(5461): 2185-2195.
doi: 10.1126/science.287.5461.2185 pmid: 10731132 |
| [3] |
Rubin GM, Yandell MD, Wortman JR, et al. Comparative genomics of the eukaryotes[J]. Science, 2000, 287(5461): 2204-2215.
pmid: 10731134 |
| [4] | 万永奇, 谢维. 生命科学与人类疾病研究的重要模型—果蝇[J]. 生命科学, 2006, 18(5): 425-429. |
| Wan YQ, Xie W. Drosophila: an important model organism for understanding basic biological and human disease mechanisms[J]. Chin Bull Life Sci, 2006, 18(5): 425-429. | |
| [5] |
Chiang HC, Hodson AC. An analytical study of population growth in Drosophila melanogaster[J]. Ecol Monogr, 1950, 20(3): 173-206.
doi: 10.2307/1948580 URL |
| [6] | Ashburner M, Thompson JN. The laboratory culture of drosophila[M]// AshburnerM, WrightT R F. The genetics and biology of drosophila. 2A. Cambridge: Academic Press, 1980: 1-81. |
| [7] | Ashburner M, Golic KG, Hawley RS. Drosophila: a laboratory handbook, 2nd ed.[M]// SciTech Book News. New York: Cold Spring Harbor Laboratory Press, 2005: 162-164. |
| [8] |
Bun-Ya M, Harashima S, Oshima Y. Putative GTP-binding protein, Gtr1, associated with the function of the Pho84 inorganic phosphate transporter in Saccharomyces cerevisiae[J]. Mol Cell Biol, 1992, 12(7): 2958-2966.
doi: 10.1128/mcb.12.7.2958-2966.1992 pmid: 1620108 |
| [9] | Panchaud N, Péli-Gulli MP, de Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1[J]. Sci Signal, 2013, 6(277): ra42. |
| [10] |
Binda M, Péli-Gulli MP, Bonfils G, et al. The Vam6 GEF controls TORC1 by activating the EGO complex[J]. Mol Cell, 2009, 35(5): 563-573.
doi: 10.1016/j.molcel.2009.06.033 pmid: 19748353 |
| [11] |
Kim J, Guan KL. mTOR as a central hub of nutrient signalling and cell growth[J]. Nat Cell Biol, 2019, 21(1): 63-71.
doi: 10.1038/s41556-018-0205-1 pmid: 30602761 |
| [12] |
Sabatini DM. Twenty-five years of mTOR: Uncovering the link from nutrients to growth[J]. PNAS, 2017, 114(45): 11818-11825.
doi: 10.1073/pnas.1716173114 pmid: 29078414 |
| [13] |
Rabanal-Ruiz Y, Otten EG, Korolchuk VI. mTORC1 as the main gateway to autophagy[J]. Essays Biochem, 2017, 61(6): 565-584.
doi: 10.1042/EBC20170027 pmid: 29233869 |
| [14] |
Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms[J]. Autophagy, 2018, 14(2): 207-215.
doi: 10.1080/15548627.2017.1378838 pmid: 28933638 |
| [15] |
Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1[J]. Science, 2008, 320(5882): 1496-1501.
doi: 10.1126/science.1157535 pmid: 18497260 |
| [16] |
Sancak Y, Bar-Peled L, Zoncu R, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids[J]. Cell, 2010, 141(2): 290-303.
doi: 10.1016/j.cell.2010.02.024 pmid: 20381137 |
| [17] |
Kim E, Goraksha-Hicks P, Li L, et al. Regulation of TORC1 by rag GTPases in nutrient response[J]. Nat Cell Biol, 2008, 10(8): 935-945.
doi: 10.1038/ncb1753 pmid: 18604198 |
| [18] |
Harrison DA, Perrimon N. Simple and efficient generation of marked clones in Drosophila[J]. Curr Biol, 1993, 3(7): 424-433.
pmid: 15335709 |
| [19] |
Gonzalez S, Rallis C. The TOR signaling pathway in spatial and temporal control of cell size and growth[J]. Front Cell Dev Biol, 2017, 5: 61.
doi: 10.3389/fcell.2017.00061 pmid: 28638821 |
| [20] |
Groth AC, Fish M, Nusse R, et al. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31[J]. Genetics, 2004, 166(4): 1775-1782.
doi: 10.1534/genetics.166.4.1775 pmid: 15126397 |
| [21] |
Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family[J]. Proc Natl Acad Sci USA, 1998, 95(10): 5505-5510.
doi: 10.1073/pnas.95.10.5505 pmid: 9576912 |
| [22] |
Fish MP, Groth AC, Calos MP, et al. Creating transgenic Drosophila by microinjecting the site-specific phiC31 integrase mRNA and a transgene-containing donor plasmid[J]. Nat Protoc, 2007, 2(10): 2325-2331.
doi: 10.1038/nprot.2007.328 |
| [23] |
Ni JQ, Markstein M, Binari R, et al. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster[J]. Nat Methods, 2008, 5(1): 49-51.
doi: 10.1038/nmeth1146 |
| [24] |
Efeyan A, Schweitzer LD, Bilate AM, et al. RagA, but not RagB, is essential for embryonic development and adult mice[J]. Dev Cell, 2014, 29(3): 321-329.
doi: 10.1016/j.devcel.2014.03.017 pmid: 24768164 |
| [25] |
Kim J, Kim E. Rag GTPase in amino acid signaling[J]. Amino Acids, 2016, 48(4): 915-928.
doi: 10.1007/s00726-016-2171-x pmid: 26781224 |
| [26] |
Efeyan A, Zoncu R, Chang S, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival[J]. Nature, 2013, 493(7434): 679-683.
doi: 10.1038/nature11745 |
| [1] | WANG Zi-yin, LIU Bing-ru, LI Zi-hao, ZHAO Xiao-yu. Characteristics of Soil Bacterial Community Structure in the Different Developmental Stages of Desert Grassland Caragana korshinskii Kom. Nebkhas [J]. Biotechnology Bulletin, 2022, 38(7): 205-214. |
| [2] | SHI Ya-qian, SHEN Ya-ru, CHEN Man-ying, HE Shu-min, LIU Yu-han, HE Tian-nan, CHEN Qing-xi, WEN Zhi-feng. Molecular Cloning and Expression Analysis of a F-box Protein Gene FnFBOX1 and Its Promoter from Fragaria nilgerrensis [J]. Biotechnology Bulletin, 2022, 38(2): 44-56. |
| [3] | LIU Shuang, YAO Jia-ni, SHEN Cong, DAI Jin-xia. Fluorescent Quantitative PCR of nifH Gene and Diversity Analysis of Nitrogen-fixing Bacteria in the Rhizosphere Soil of Caragana spp. of Desert Grassland [J]. Biotechnology Bulletin, 2022, 38(12): 252-262. |
| [4] | HONG Ge-riqiqige, WANG Yan-fei, GAO Xian-ling, PANG Cai-xia, SHANG Xiao-rui, LI Guo-jing, WANG Rui-gang. Cloning of CiNAC038 Gene Promoter in Caragana intermedia and Profiling of Its Responses to Hormones [J]. Biotechnology Bulletin, 2020, 36(7): 55-61. |
| [5] | ZHAO Xiao-xia, NIU Shi-quan, WEN Na, SU Feng-feng. Screening and Identification of Biocontrol Bacillus sp. Against Astragalus Root Rot and Its Pot Experiment [J]. Biotechnology Bulletin, 2019, 35(9): 107-111. |
| [6] | ZHANG Meng, LIU Guo-lin, LI Xiang-long, CHEN Yong-hong, BAI Ling-rong, LUO Fang, LI Ya-chao, TAO Jin-zhong. Effect of Adding Hawthorn and Astragalus Mixtures on the Plasma Metabolome of Perinatal Dairy Cows [J]. Biotechnology Bulletin, 2019, 35(8): 127-137. |
| [7] | YANG Fei-yun, BAI Jie, LIU Kun, WANG Rui-gang. Increase of Flavonoids Content in Arabidopsis Heterologous Expressing CiCHI Gene [J]. Biotechnology Bulletin, 2019, 35(11): 39-45. |
| [8] | ZHANG Teng-guo, SHI Zhong-fei, KOU Ming-gang, ZHENG Sheng, WANG Juan. Cloning and Expression Analysis of CkP5CS Gene in Caragana korshinskii [J]. Biotechnology Bulletin, 2018, 34(9): 209-218. |
| [9] | LI Min,QU Jie,LI Meng-jie,HU Xiao-long. Function of Osa in the Differentiation of Drosophila Ovary Germline Stem Cell [J]. Biotechnology Bulletin, 2017, 33(9): 139-144. |
| [10] | CHEN Zhe, HUANG Jing, ZHAO Jia, LIANG Hong. Research Advance on the Red Stele Root Rot of Strawberry [J]. Biotechnology Bulletin, 2017, 33(3): 37-44. |
| [11] | WANG Guang-xia, YANG Qi, WANG Rui-gang, LI Guo-jing. Systematical Identification and Primer Screening of EST-SSR Marker in Transcriptome of Caragana intermedia [J]. Biotechnology Bulletin, 2016, 32(2): 178-184. |
| [12] | Liu Lili Wang Jian Wang Haisheng Yu Kaimin Li Guochao Yan Yanchun . Foundation of Platform for Zebrafish Transgenic Technology [J]. Biotechnology Bulletin, 2013, 0(10): 120-126. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||