








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 137-147.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0138
Previous Articles Next Articles
ZHAO Si-jia1( ), WANG Xiao-lu2, SUN Ji-lu1, TIAN Jian2(
), WANG Xiao-lu2, SUN Ji-lu1, TIAN Jian2( ), ZHANG Jie2(
), ZHANG Jie2( )
)
Received:2023-02-19
Online:2023-08-26
Published:2023-09-05
Contact:
TIAN Jian, ZHANG Jie
E-mail:sijia_zhao@163.com;tianjian@caas.cn;zhangjie09@caas.cn
ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering[J]. Biotechnology Bulletin, 2023, 39(8): 137-147.
| 基因Gene name | 功能Function | 来源Source | 蛋白分子量Protein molecular weight/kD |
|---|---|---|---|
| pyp1 | 糖醇磷酸酶基因Sugar alcohol phosphatase gene | S. cerevisiae | 30.7 |
| yidA | 去磷酸化酶基因Dphosphorylase gene | E. coli MG1655 | 32.9 |
| err1 | 赤藓糖还原酶基因Erythrose reductase gene | T. reesei | 39.5 |
| er(NADH) | 赤藓糖还原酶基因Erythrose reductase gene | Y. lipolytica | 39.4 |
| er(NADPH) | 赤藓糖还原酶基因Erythrose reductase gene | C. magnoliae JH110 | 34.6 |
| er1 | 赤藓糖还原酶基因Erythrose reductase gene | M. megachiliensis | 39.6 |
| er3 | 赤藓糖还原酶基因Erythrose reductase gene | M. megachiliensis | 39.8 |
| ER10 | 赤藓糖还原酶基因Erythrose reductase gene | Y. lipolytica CLIB122 | 39.1 |
| ER25 | 赤藓糖还原酶基因Erythrose reductase gene | Y. lipolytica CLIB122 | 38.2 |
| JX885 | 赤藓糖还原酶基因Erythrose reductase gene | Y. lipolytica | 31.4 |
| rpe1 | 差向异构酶基因Epimerase gene | S. cerevisiae | 29.2 |
Table 1 Exogenous genes involved in this study
| 基因Gene name | 功能Function | 来源Source | 蛋白分子量Protein molecular weight/kD |
|---|---|---|---|
| pyp1 | 糖醇磷酸酶基因Sugar alcohol phosphatase gene | S. cerevisiae | 30.7 |
| yidA | 去磷酸化酶基因Dphosphorylase gene | E. coli MG1655 | 32.9 |
| err1 | 赤藓糖还原酶基因Erythrose reductase gene | T. reesei | 39.5 |
| er(NADH) | 赤藓糖还原酶基因Erythrose reductase gene | Y. lipolytica | 39.4 |
| er(NADPH) | 赤藓糖还原酶基因Erythrose reductase gene | C. magnoliae JH110 | 34.6 |
| er1 | 赤藓糖还原酶基因Erythrose reductase gene | M. megachiliensis | 39.6 |
| er3 | 赤藓糖还原酶基因Erythrose reductase gene | M. megachiliensis | 39.8 |
| ER10 | 赤藓糖还原酶基因Erythrose reductase gene | Y. lipolytica CLIB122 | 39.1 |
| ER25 | 赤藓糖还原酶基因Erythrose reductase gene | Y. lipolytica CLIB122 | 38.2 |
| JX885 | 赤藓糖还原酶基因Erythrose reductase gene | Y. lipolytica | 31.4 |
| rpe1 | 差向异构酶基因Epimerase gene | S. cerevisiae | 29.2 |
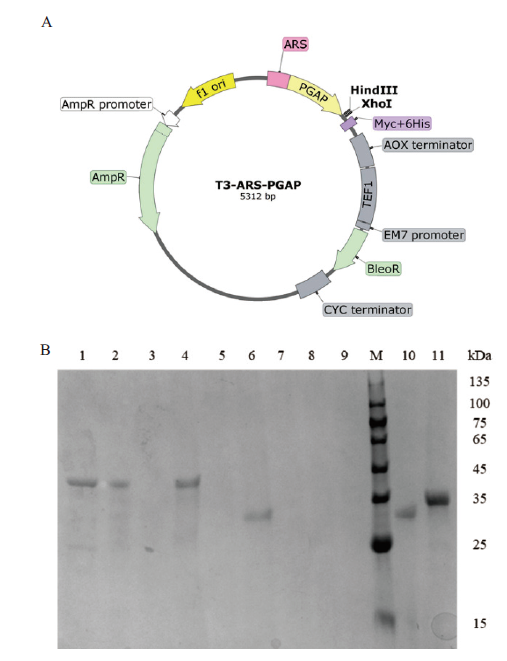
Fig. 4 Expressions of exogenous genes in P. pastoris A: T-ARS-PGAP plasmid map; B: SDS-PAGE detection of the expression of exogenous genes. 1-11: err1, er(NADH), er(NADPH), er, er3, pyp1, ER10, ER25, JX885, rpe1, yidA
| 化合物名称Compound name | 最高浓度Maximum concentration/(g·L-1) |
|---|---|
| 赤藓糖醇 Erythritol | 0 |
| 阿拉伯糖醇 Arabitol | 1.4 |
| 核糖醇 Ribitol | 5.6 |
Table 2 Products of high-density fermentation of mutant C5
| 化合物名称Compound name | 最高浓度Maximum concentration/(g·L-1) |
|---|---|
| 赤藓糖醇 Erythritol | 0 |
| 阿拉伯糖醇 Arabitol | 1.4 |
| 核糖醇 Ribitol | 5.6 |
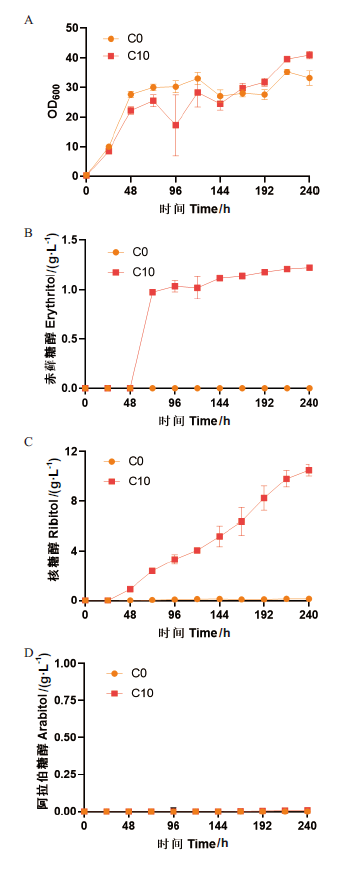
Fig. 6 Phenotype verification of P. pastoris C0 and C10 strains A: Growth curve. B: Production of erythritol. C: Production of ribitol. D: Production of arabitol
| 化合物名称 Compound name | 最高浓度Maximum concentration/(g·L-1) | |
|---|---|---|
| C11 | C12 | |
| 赤藓糖醇 Erythritol | 1.2 | 1.2 |
| 核糖醇 Ribitol | 1.6 | 4.0 |
| 阿拉伯糖醇 Arabitol | 4.9 | 5.1 |
Table 3 Products of shake-flask fermentation of mutant C11 and C12
| 化合物名称 Compound name | 最高浓度Maximum concentration/(g·L-1) | |
|---|---|---|
| C11 | C12 | |
| 赤藓糖醇 Erythritol | 1.2 | 1.2 |
| 核糖醇 Ribitol | 1.6 | 4.0 |
| 阿拉伯糖醇 Arabitol | 4.9 | 5.1 |
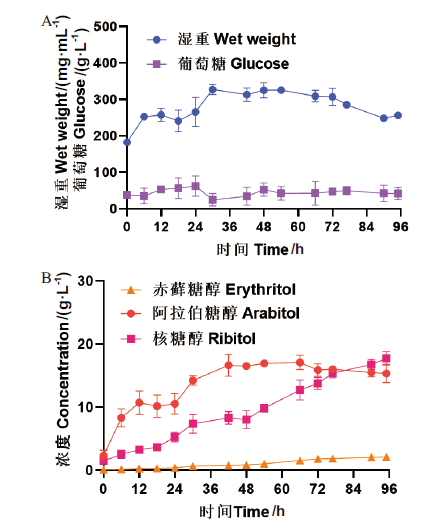
Fig. 8 High-cell-density fermentation with strain P. pastoris C11 A: Wet weight and glucose concentration. B: Production of erythritol, ribitol and arabitol
| [1] |
Hijosa-Valsero M, Garita-Cambronero J, Paniagua-García AI, et al. By-products of sugar factories and wineries as feedstocks for erythritol generation[J]. Food Bioprod Process, 2021, 126: 345-355.
doi: 10.1016/j.fbp.2021.02.001 URL |
| [2] | 中国食品科学技术学会. 赤藓糖醇的科学共识[J]. 中国食品学报, 2022, 22(12): 405-412. |
| Chinese Institute of Food Science and Technology. Scientific consensus on erythritol[J]. J Chin Inst Food Sci Technol, 2022, 22(12): 405-412. | |
| [3] |
Martău GA, Coman V, Vodnar DC. Recent advances in the biotechnological production of erythritol and mannitol[J]. Crit Rev Biotechnol, 2020, 40(5): 608-622.
doi: 10.1080/07388551.2020.1751057 pmid: 32299245 |
| [4] | 邱学良. 产赤藓糖醇亚罗解脂酵母的耐热机制分析及组合策略改造[D]. 无锡: 江南大学, 2020. |
| Qiu XL. Thermotolerance mechanism analysis and combination strategy transformation of erythritol producing Yarrowia lipolytica[D]. Wuxi: Jiangnan University, 2020. | |
| [5] |
den Hartog GJM, Boots AW, Adam-Perrot A, et al. Erythritol is a sweet antioxidant[J]. Nutrition, 2010, 26(4): 449-458.
doi: 10.1016/j.nut.2009.05.004 pmid: 19632091 |
| [6] | 张妍, 张丽, 李宝磊, 等. 赤藓糖醇的国内外研究进展[J]. 饮料工业, 2022, 25(5): 76-79. |
| Zhang Y, Zhang L, Li BL, et al. Research progress of erythritol at home and abroad[J]. Beverage Ind, 2022, 25(5): 76-79. | |
| [7] | 中华人民共和国工业和信息化部. 口腔清洁护理用品牙膏中赤藓糖醇含量的测定高效液相色谱法: QB/T 5408—2019[S]. 北京: 中国轻工业出版社, 2019. |
| Ministry of Industry and Information of the People's Republic of China. Oral hygiene care products determination of erythritol content in toothpaste high performance liquid chromatography: QB/T 5408—2019[S]. Beijing: China Light Industry Press, 2019. | |
| [8] |
Liang PX, Cao MF, Li J, et al. Expanding sugar alcohol industry: Microbial production of sugar alcohols and associated chemocatalytic derivatives[J]. Biotechnol Adv, 2023, 64: 108105.
doi: 10.1016/j.biotechadv.2023.108105 URL |
| [9] | 隋松森, 王松江, 郭传庄, 等. 微生物发酵法产赤藓糖醇的研究进展[J]. 中国食品添加剂, 2021, 32(6): 125-131. |
| Sui SS, Wang SJ, Guo CZ, et al. Research progress in erythritol production by microbial fermentation[J]. China Food Addit, 2021, 32(6): 125-131. | |
| [10] | Stapley JA, Genders DJ, Atherton DM, et al. Methods for the electrolytic production of erythrose or erythritol: United States: US7955489[P]. 2011-06-07. |
| [11] |
Richter H, Vlad D, Unden G. Significance of pantothenate for glucose fermentation by Oenococcus oeni and for suppression of the erythritol and acetate production[J]. Arch Microbiol, 2001, 175(1): 26-31.
pmid: 11271417 |
| [12] |
Gänzle MG, Vermeulen N, Vogel RF. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough[J]. Food Microbiol, 2007, 24(2): 128-138.
pmid: 17008155 |
| [13] |
Veiga-da-Cunha M, Santos H, Van Schaftingen E. Pathway and regulation of erythritol formation in Leuconostoc oenos[J]. J Bacteriol, 1993, 175(13): 3941-3948.
pmid: 8391532 |
| [14] |
Khatape AB, Dastager SG, Rangaswamy V. An overview of erythritol production by yeast strains[J]. FEMS Microbiol Lett, 2022, 369(1): fnac107.
doi: 10.1093/femsle/fnac107 URL |
| [15] |
Rakicka-Pustułka M, Mirończuk AM, Celińska E, et al. Scale-up of the erythritol production technology - Process simulation and techno-economic analysis[J]. J Clean Prod, 2020, 257: 120533.
doi: 10.1016/j.jclepro.2020.120533 URL |
| [16] |
Deshpande MS, Kulkarni PP, Kumbhar PS, et al. Erythritol production from sugar based feedstocks by Moniliella pollinis using lysate of recycled cells as nutrients source[J]. Process Biochem, 2022, 112: 45-52.
doi: 10.1016/j.procbio.2021.11.020 URL |
| [17] |
Yang SL, Pan XW, Wang Q, et al. Enhancing erythritol production from crude glycerol in a wild-type Yarrowia lipolytica by metabolic engineering[J]. Front Microbiol, 2022, 13: 1054243.
doi: 10.3389/fmicb.2022.1054243 URL |
| [18] |
Li LZ, Yang TY, Guo WQ, et al. Construction of an efficient mutant strain of Trichosporonoides oedocephalis with HOG1 gene deletion for production of erythritol[J]. J Microbiol Biotechnol, 2016, 26(4): 700-709.
doi: 10.4014/jmb.1510.10049 URL |
| [19] |
Gómez-Ramírez IV, Corrales-García LL, Possani LD, et al. Expression in Pichia pastoris of human antibody fragments that neutralize venoms of Mexican scorpions[J]. Toxicon, 2023, 223: 107012.
doi: 10.1016/j.toxicon.2022.107012 URL |
| [20] |
Guo F, Dai ZX, Peng WF, et al. Metabolic engineering of Pichia pastoris for malic acid production from methanol[J]. Biotechnol Bioeng, 2021, 118(1): 357-371.
doi: 10.1002/bit.v118.1 URL |
| [21] | Cai P, Wu XY, Deng J, et al. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris[J]. Proc Natl Acad Sci USA, 2022, 119(29): e2201711119. |
| [22] |
Zhang QQ, Wang XL, Luo HY, et al. Metabolic engineering of Pichia pastoris for myo-inositol production by dynamic regulation of central metabolism[J]. Microb Cell Fact, 2022, 21(1): 112.
doi: 10.1186/s12934-022-01837-x |
| [23] |
Abdel-Mawgoud AM, Markham KA, Palmer CM, et al. Metabolic engineering in the host Yarrowia lipolytica[J]. Metab Eng, 2018, 50: 192-208.
doi: S1096-7176(18)30273-8 pmid: 30056205 |
| [24] |
Kobayashi Y, Yoshida J, Iwata H, et al. Gene expression and function involved in polyol biosynthesis of Trichosporonoides megachiliensis under hyper-osmotic stress[J]. J Biosci Bioeng, 2013, 115(6): 645-650.
doi: 10.1016/j.jbiosc.2012.12.004 pmid: 23294575 |
| [25] |
Lee DH, Lee YJ, Ryu YW, et al. Molecular cloning and biochemical characterization of a novel erythrose reductase from Candida magnoliae JH110[J]. Microb Cell Fact, 2010, 9: 43.
doi: 10.1186/1475-2859-9-43 |
| [26] |
Carly F, Vandermies M, Telek S, et al. Enhancing erythritol productivity in Yarrowia lipolytica using metabolic engineering[J]. Metab Eng, 2017, 42: 19-24.
doi: 10.1016/j.ymben.2017.05.002 URL |
| [27] |
Jovanović B, Mach RL, Mach-Aigner AR. Characterization of erythrose reductases from filamentous fungi[J]. AMB Express, 2013, 3(1): 43.
doi: 10.1186/2191-0855-3-43 pmid: 23924507 |
| [28] |
Janek T, Dobrowolski A, Biegalska A, et al. Characterization of erythrose reductase from Yarrowia lipolytica and its influence on erythritol synthesis[J]. Microb Cell Fact, 2017, 16(1): 118.
doi: 10.1186/s12934-017-0733-6 URL |
| [29] |
Shenoy A, Yalamanchili S, Davis AR, et al. Expression and display of glycoengineered antibodies and antibody fragments with an engineered yeast strain[J]. Antibodies, 2021, 10(4): 38.
doi: 10.3390/antib10040038 URL |
| [30] |
De Brabander P, Uitterhaegen E, Delmulle T, et al. Challenges and progress towards industrial recombinant protein production in yeasts: a review[J]. Biotechnol Adv, 2023, 64: 108121.
doi: 10.1016/j.biotechadv.2023.108121 URL |
| [31] |
Ávila-Fernández Á, Montiel S, Rodríguez-Alegría ME, et al. Simultaneous enzyme production, Levan-type FOS synthesis and sugar by-products elimination using a recombinant Pichia pastoris strain expressing a levansucrase-endolevanase fusion enzyme[J]. Microb Cell Fact, 2023, 22(1): 18.
doi: 10.1186/s12934-022-02009-7 pmid: 36703199 |
| [32] |
Inokuma K, Miyamoto S, Morinaga K, et al. Direct production of 4-hydroxybenzoic acid from cellulose using cellulase-displaying Pichia pastoris[J]. Biotechnol Bioeng, 2023, 120(4): 1097-1107.
doi: 10.1002/bit.v120.4 URL |
| [33] |
Chen SL, Liu TS, Zhang WG, et al. Cofactor engineering for efficient production of α-farnesene by rational modification of NADPH and ATP regeneration pathway in Pichia pastoris[J]. Int J Mol Sci, 2023, 24(2): 1767.
doi: 10.3390/ijms24021767 URL |
| [34] |
Hijosa-Valsero M, Paniagua-García AI, Díez-Antolínez R. Cell immobilization for erythritol production[J]. J Fungi, 2022, 8(12): 1286.
doi: 10.3390/jof8121286 URL |
| [1] | WU Qiao-yin, SHI You-zhi, LI Lin-lin, PENG Zheng, TAN Zai-yu, LIU Li-ping, ZHANG Juan, PAN Yong. In Situ Screening of Carotenoid Degrading Strains and the Application in Improving Quality and Aroma of Cigar [J]. Biotechnology Bulletin, 2023, 39(9): 192-201. |
| [2] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [3] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [4] | XU Fa-di, XU Kang, SUN Dong-ming, LI Meng-lei, ZHAO Jian-zhi, BAO Xiao-ming. Research Progress in Second-generation Fuel Ethanol Technology Based on Poplar(Populus sp.) [J]. Biotechnology Bulletin, 2023, 39(9): 27-39. |
| [5] | ZHANG Yue-yi, LAN She-yi, PEI Hai-run, FENG Di. Process Optimization of Multi-strain Fermented Oat Bran and Hair Efficacy Evaluation [J]. Biotechnology Bulletin, 2023, 39(9): 58-70. |
| [6] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [7] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [8] | DONG Cong, GAO Qing-hua, WANG Yue, LUO Tong-yang, WANG Qing-qing. Increasing the Expression of FAD-dependent Glucose Dehydrogenase by Recombinant Pichia pastoris Using a Combined Strategy [J]. Biotechnology Bulletin, 2023, 39(6): 316-324. |
| [9] | YU Hui-li, LI Ai-tao. Application of Cytochrome P450 in the Biosynthesis of Flavors and Fragrances [J]. Biotechnology Bulletin, 2023, 39(4): 24-37. |
| [10] | LI Xin-yue, ZHOU Ming-hai, FAN Ya-chao, LIAO Sha, ZHANG Feng-li, LIU Chen-guang, SUN Yue, ZHANG Lin, ZHAO Xin-qing. Research Progress in the Improvement of Microbial Strain Tolerance and Efficiency of Biological Manufacturing Based on Transporter Engineering [J]. Biotechnology Bulletin, 2023, 39(11): 123-136. |
| [11] | CHE Yong-mei, LIU Guang-chao, GUO Yan-ping, YE Qing, ZHAO Fang-gui, LIU Xin. Preparation of Compound Halotolerant Bioinoculant and Study on Its Growth-promoting Effect [J]. Biotechnology Bulletin, 2023, 39(11): 217-225. |
| [12] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [13] | REN Hai-wei, SUN Yi-fan, REN Yu-wei, GUO Xiao-peng, PAN Li-chao, ZHANG Bing-yun, LI Jin-ping. Research Progress of Silage Additives Based on Bibliometrics [J]. Biotechnology Bulletin, 2022, 38(8): 261-274. |
| [14] | WANG Xin-guang, TIAN Lei, WANG En-ze, ZHONG Cheng, TIAN Chun-jie. Construction of Microbial Consortium for Efficient Degradation of Corn Straw and Evaluation of Its Degradation Effect [J]. Biotechnology Bulletin, 2022, 38(4): 217-229. |
| [15] | WANG Yue, GAO Qing-hua, DONG Cong, LUO Tong-yang, WANG Qing-qing. Expression of Pyranose Oxidase with Optimized Codon in Pichia pastoris [J]. Biotechnology Bulletin, 2022, 38(4): 269-277. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||