








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 126-136.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1530
Previous Articles Next Articles
SHA Shan-shan( ), DONG Shi-rong, YANG Yu-ju(
), DONG Shi-rong, YANG Yu-ju( )
)
Received:2022-12-12
Online:2023-08-26
Published:2023-09-05
Contact:
YANG Yu-ju
E-mail:18045187642@163.com;sh_yangyj@hrbu.edu.cn
SHA Shan-shan, DONG Shi-rong, YANG Yu-ju. Research Progress in Gut Microbiota and Metabolites Regulating Host Intestinal Immunity[J]. Biotechnology Bulletin, 2023, 39(8): 126-136.
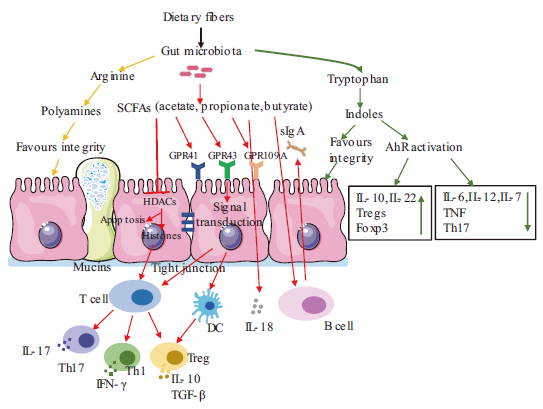
Fig. 1 Regulation of gut microbiota and metabolites on intestinal immune system The narrow arrows indicate promotions, flat-head arrows indicate inhibitions. SCFAs: Short chain fatty acid. HDACs: Histone deacetylase. IL: Interleukin. Th: Helper T cells. Treg: Regulatory T cell. TGF-β: Transforming growth factor beta. GPR: G-protein-coupled receptors. sIgA: Secretory immunoglobulin A. AhR activation: Aromatic hydrocarbon receptor activation. Foxp3: Fork-headed transcription factor p3. TNF: Tumor necrosis factor. DC: Dendritic cell. IFN-γ: Gamma interferon
| [1] | 李星, 曹振辉, 林秋叶, 等. 肠道微生物及其代谢产物对动物免疫机能的影响[J]. 动物营养学报, 2019, 31(2): 553-559. |
| Li X, Cao ZH, Lin QY, et al. Effects of gut microbiota and its metabolites on animal immune function[J]. Chin J Animal Nutr, 2019, 31(2): 553-559. | |
| [2] | Cummings JH, Antoine JM, Azpiroz F, et al. PASSCLAIM1? Gut health and immunity[J]. Eur J Nutr, 2004, 43(S2): ii118-ii173. |
| [3] |
Turner JR. Intestinal mucosal barrier function in health and disease[J]. Nat Rev Immunol, 2009, 9(11): 799-809.
doi: 10.1038/nri2653 pmid: 19855405 |
| [4] | 金磊, 王立志. 肠道微生物与宿主免疫关系研究进展[J]. 现代畜牧兽医, 2018(9): 52-58. |
| Jin L, Wang LZ. Research progress on the immunological relationship between intestinal microorganism and host[J]. Mod J Animal Husb Vet Med, 2018(9): 52-58. | |
| [5] |
Burgueño JF, Abreu MT. Epithelial Toll-like receptors and their role in gut homeostasis and disease[J]. Nat Rev Gastroenterol Hepatol, 2020, 17(5): 263-278.
doi: 10.1038/s41575-019-0261-4 pmid: 32103203 |
| [6] |
Liu YH, Ajami NJ, El-Serag HB, et al. Dietary quality and the colonic mucosa-associated gut microbiome in humans[J]. Am J Clin Nutr, 2019, 110(3): 701-712.
doi: 10.1093/ajcn/nqz139 pmid: 31291462 |
| [7] |
Suzuki T. Regulation of the intestinal barrier by nutrients: The role of tight junctions[J]. Anim Sci J, 2020, 91(1): e13357.
doi: 10.1111/asj.v91.1 URL |
| [8] |
Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target?[J]. Nat Rev Gastroenterol Hepatol, 2017, 14(1): 9-21.
doi: 10.1038/nrgastro.2016.169 pmid: 27848962 |
| [9] |
McLoughlin RF, Berthon BS, Jensen ME, et al. Short-chain fatty acids, prebiotics, synbiotics, and systemic inflammation: a systematic review and meta-analysis[J]. Am J Clin Nutr, 2017, 106(3): 930-945.
doi: 10.3945/ajcn.117.156265 pmid: 28793992 |
| [10] |
Uematsu S, Fujimoto K, Jang MH, et al. Regulation of humoral and cellular gut immunity by Lamina propria dendritic cells expressing Toll-like receptor 5[J]. Nat Immunol, 2008, 9(7): 769-776.
doi: 10.1038/ni.1622 |
| [11] |
Tong TJ, Qi YJ, Bussiere LD, et al. Transport of artificial virus-like nanocarriers through intestinal monolayers via microfold cells[J]. Nanoscale, 2020, 12(30): 16339-16347.
doi: 10.1039/D0NR03680C URL |
| [12] |
Caricilli AM, Castoldi A, Câmara NOS. Intestinal barrier: a gentlemen's agreement between microbiota and immunity[J]. World J Gastrointest Pathophysiol, 2014, 5(1): 18-32.
doi: 10.4291/wjgp.v5.i1.18 pmid: 24891972 |
| [13] |
Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease[J]. Nat Rev Immunol, 2013, 13(5): 321-335.
doi: 10.1038/nri3430 pmid: 23618829 |
| [14] | Milani C, Duranti S, Bottacini F, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota[J]. Microbiol Mol Biol Rev, 2017, 81(4): e00036-e00017. |
| [15] | Zou SM, Fang LK, Lee MH. Dysbiosis of gut microbiota in promoting the development of colorectal cancer[J]. Gastroenterol Rep(Oxf), 2018, 6(1): 1-12. |
| [16] |
Mao JD. Lactobacillus rhamnosus GG attenuates lipopolysaccharide-induced inflammation and barrier dysfunction by regulating MAPK/NF-ĸB signaling and modulating metabolome in the piglet intestine[J]. J Nutr, 2020, 150(5): 1313-1323.
doi: 10.1093/jn/nxaa009 URL |
| [17] |
Brink LR, Mercer KE, Piccolo BD, et al. Neonatal diet alters fecal microbiota and metabolome profiles at different ages in infants fed breast milk or formula[J]. Am J Clin Nutr, 2020, 111(6): 1190-1202.
doi: 10.1093/ajcn/nqaa076 pmid: 32330237 |
| [18] |
Sommer F, Bäckhed F. The gut microbiota—Masters of host development and physiology[J]. Nat Rev Microbiol, 2013, 11(4): 227-238.
doi: 10.1038/nrmicro2974 pmid: 23435359 |
| [19] | 郭宏伟, 张妮妮, 张伟, 等. 抗生素诱导的菌群紊乱对幼鼠结肠黏膜屏障及免疫反应的影响[J]. 中华实用儿科临床杂志, 2019, 34(7): 505-509. |
| Guo HW, Zhang NN, Zhang W, et al. Effect of antibiotic-induced microbiota dysbiosis on colonic mucosal barrier and immune response in juvenile mice[J]. Chin J Appl Clin Pediatr, 2019, 34(7): 505-509. | |
| [20] |
Ge XL, Ding C, Zhao W, et al. Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility[J]. J Transl Med, 2017, 15(1): 13.
doi: 10.1186/s12967-016-1105-4 pmid: 28086815 |
| [21] |
Franzosa EA, Hsu T, Sirota-Madi A, et al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling[J]. Nat Rev Microbiol, 2015, 13(6): 360-372.
doi: 10.1038/nrmicro3451 pmid: 25915636 |
| [22] |
Zuñiga C, Zaramela L, Zengler K. Elucidation of complexity and prediction of interactions in microbial communities[J]. Microb Biotechnol, 2017, 10(6): 1500-1522.
doi: 10.1111/1751-7915.12855 pmid: 28925555 |
| [23] | 侯璐文, 吴长新, 秦雪梅, 等. 肠道微生物功能宏基因组学与代谢组学关联分析方法研究进展[J]. 微生物学报, 2019, 59(9): 1813-1822. |
| Hou LW, Wu CX, Qin XM, et al. Research progress on correlation analysis methods of functional metagenomics and metabonomics of intestinal microorganisms[J]. Acta Microbiol Sin, 2019, 59(9): 1813-1822. | |
| [24] |
Zheng YP, Ran Y, Zhang HX, et al. The microbiome in autoimmune liver diseases: metagenomic and metabolomic changes[J]. Front Physiol, 2021, 12: 715852.
doi: 10.3389/fphys.2021.715852 URL |
| [25] |
Wang YL, Yin YS, Chen X, et al. Induction of intestinal Th17 cells by flagellins from segmented filamentous bacteria[J]. Front Immunol, 2019, 10: 2750.
doi: 10.3389/fimmu.2019.02750 pmid: 31824516 |
| [26] |
Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species[J]. Science, 2011, 331(6015): 337-341.
doi: 10.1126/science.1198469 pmid: 21205640 |
| [27] |
Wong SH. Zhao LY, Zhang X, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice[J]. Gastroenterology, 2017, 153(6): 1621-1633.
doi: 10.1053/j.gastro.2017.08.022 URL |
| [28] |
Thiele Orberg E, Fan H, Tam AJ, et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis[J]. Mucosal Immunol, 2017, 10(2): 421-433.
doi: 10.1038/mi.2016.53 pmid: 27301879 |
| [29] |
Coleman OI, Lobner EM, Bierwirth S, et al. Activated ATF6 induces intestinal dysbiosis and innate immune response to promote colorectal tumorigenesis[J]. Gastroenterology, 2018, 155(5): 1539-1552.
doi: S0016-5085(18)34816-9 pmid: 30063920 |
| [30] |
Jiao YH, Wu L, Huntington ND, et al. Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases[J]. Front Immunol, 2020, 11: 282.
doi: 10.3389/fimmu.2020.00282 pmid: 32153586 |
| [31] |
Chida S, Sakamoto M, Takino T, et al. Changes in immune system and intestinal bacteria of cows during the transition period[J]. Vet Anim Sci, 2021, 14: 100222.
doi: 10.1016/j.vas.2021.100222 URL |
| [32] |
Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens[J]. Nat Rev Immunol, 2013, 13(11): 790-801.
doi: 10.1038/nri3535 pmid: 24096337 |
| [33] |
Schwartz DJ, Langdon AE, Dantas G. Understanding the impact of antibiotic perturbation on the human microbiome[J]. Genome Med, 2020, 12(1): 82.
doi: 10.1186/s13073-020-00782-x pmid: 32988391 |
| [34] |
Zimmermann P, Curtis N. The effect of antibiotics on the composition of the intestinal microbiota - a systematic review[J]. J Infect, 2019, 79(6): 471-489.
doi: S0163-4453(19)30318-4 pmid: 31629863 |
| [35] |
Wu HG, Chen Q, Liu JN, et al. Microbiome analysis reveals gut microbiota alteration in mice with the effect of matrine[J]. Microb Pathog, 2021, 156: 104926.
doi: 10.1016/j.micpath.2021.104926 URL |
| [36] |
Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity[J]. Nat Rev Immunol, 2016, 16(6): 341-352.
doi: 10.1038/nri.2016.42 pmid: 27231050 |
| [37] |
Koh A, De Vadder F, Kovatcheva-Datchary P, et al. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites[J]. Cell, 2016, 165(6): 1332-1345.
doi: S0092-8674(16)30592-X pmid: 27259147 |
| [38] |
Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer[J]. Nat Rev Microbiol, 2014, 12(10): 661-672.
doi: 10.1038/nrmicro3344 pmid: 25198138 |
| [39] |
Reichardt N, Duncan SH, Young P, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota[J]. ISME J, 2014, 8(6): 1323-1335.
doi: 10.1038/ismej.2014.14 pmid: 24553467 |
| [40] |
Flint HJ, Duncan SH, Scott KP, et al. Links between diet, gut microbiota composition and gut metabolism[J]. Proc Nutr Soc, 2015, 74(1): 13-22.
doi: 10.1017/S0029665114001463 pmid: 25268552 |
| [41] |
Vinolo MAR, Rodrigues HG, Hatanaka E, et al. Short-chain fatty acids stimulate the migration of neutrophils to inflammatory sites[J]. Clin Sci(Lond), 2009, 117(9): 331-338.
doi: 10.1042/CS20080642 URL |
| [42] |
Vinolo MAR, Hatanaka E, Lambertucci RH, et al. Effects of short chain fatty acids on effector mechanisms of neutrophils[J]. Cell Biochem Funct, 2009, 27(1): 48-55.
doi: 10.1002/cbf.1533 pmid: 19107872 |
| [43] |
Vinolo MAR, Rodrigues HG, Hatanaka E, et al. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils[J]. J Nutr Biochem, 2011, 22(9): 849-855.
doi: 10.1016/j.jnutbio.2010.07.009 pmid: 21167700 |
| [44] |
Nastasi C, Candela M, Bonefeld CM, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells[J]. Sci Rep, 2015, 5: 16148.
doi: 10.1038/srep16148 pmid: 26541096 |
| [45] |
Li YJ, Chen X, Kwan TK, et al. Dietary fiber protects against diabetic nephropathy through short-chain fatty acid-mediated activation of G protein-coupled receptors GPR43 and GPR109A[J]. J Am Soc Nephrol, 2020, 31(6): 1267-1281.
doi: 10.1681/ASN.2019101029 pmid: 32358041 |
| [46] | Grabarska A, Dmoszyńska-Graniczka M, Nowosadzka E, et al. Histone deacetylase inhibitors - molecular mechanisms of actions and clinical applications[J]. Postepy Hig Med Dosw(Online), 2013, 67: 722-735. |
| [47] |
Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis[J]. Science, 2013, 341(6145): 569-573.
doi: 10.1126/science.1241165 pmid: 23828891 |
| [48] |
Tao R, de Zoeten EF, Ozkaynak E, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells[J]. Nat Med, 2007, 13(11):1299-1307.
doi: 10.1038/nm1652 pmid: 17922010 |
| [49] |
Chang PV, Hao LM, Offermanns S, et al. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition[J]. Proc Natl Acad Sci USA, 2014, 111(6): 2247-2252.
doi: 10.1073/pnas.1322269111 pmid: 24390544 |
| [50] |
Singh N, Thangaraju M, Prasad PD, et al. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter(Slc5a8)-dependent inhibition of histone deacetylases[J]. J Biol Chem, 2010, 285(36): 27601-27608.
doi: 10.1074/jbc.M110.102947 URL |
| [51] |
Hino S, Mizushima T, Kaneko K, et al. Mucin-derived O-glycans act as endogenous fiber and sustain mucosal immune homeostasis via short-chain fatty acid production in rat cecum[J]. J Nutr, 2020, 150(10): 2656-2665.
doi: 10.1093/jn/nxaa097 pmid: 32286621 |
| [52] |
Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis[J]. Immunity, 2014, 40(1): 128-139.
doi: 10.1016/j.immuni.2013.12.007 pmid: 24412617 |
| [53] |
Yang WJ, Yu TM, Huang XS, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity[J]. Nat Commun, 2020, 11(1): 4457.
doi: 10.1038/s41467-020-18262-6 pmid: 32901017 |
| [54] |
Park J, Kim M, Kang SG, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway[J]. Mucosal Immunol, 2015, 8(1): 80-93.
doi: 10.1038/mi.2014.44 pmid: 24917457 |
| [55] |
Li Y, Innocentin S, Withers DR, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation[J]. Cell, 2011, 147(3): 629-640.
doi: 10.1016/j.cell.2011.09.025 pmid: 21999944 |
| [56] |
Natividad JM, Agus A, Planchais J, et al. Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome[J]. Cell Metab, 2018, 28(5): 737-749.
doi: S1550-4131(18)30444-3 pmid: 30057068 |
| [57] |
Murray IA, Nichols RG, Zhang LM, et al. Expression of the aryl hydrocarbon receptor contributes to the establishment of intestinal microbial community structure in mice[J]. Sci Rep, 2016, 6: 33969.
doi: 10.1038/srep33969 pmid: 27659481 |
| [58] |
Zhang LM, Nichols RG, Correll J, et al. Persistent organic pollutants modify gut microbiota-host metabolic homeostasis in mice through aryl hydrocarbon receptor activation[J]. Environ Health Perspect, 2015, 123(7): 679-688.
doi: 10.1289/ehp.1409055 URL |
| [59] |
Lamas B, Natividad JM, Sokol H. Aryl hydrocarbon receptor and intestinal immunity[J]. Mucosal Immunol, 2018, 11(4): 1024-1038.
doi: 10.1038/s41385-018-0019-2 pmid: 29626198 |
| [60] |
Yang F, DeLuca JAA, Menon R, et al. Effect of diet and intestinal AhR expression on fecal microbiome and metabolomic profiles[J]. Microb Cell Fact, 2020, 19(1): 219.
doi: 10.1186/s12934-020-01463-5 pmid: 33256731 |
| [61] |
Hubbard TD, Murray IA, Bisson WH, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles[J]. Sci Rep, 2015, 5: 12689.
doi: 10.1038/srep12689 pmid: 26235394 |
| [62] |
Sun M, Ma N, He T, et al. Tryptophan(Trp)modulates gut homeostasis via aryl hydrocarbon receptor(AhR)[J]. Crit Rev Food Sci Nutr, 2020, 60(10): 1760-1768.
doi: 10.1080/10408398.2019.1598334 URL |
| [63] |
Qiu JX, Guo Z, Chen L, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora[J]. Immunity, 2013, 39(2): 386-399.
doi: 10.1016/j.immuni.2013.08.002 pmid: 23954130 |
| [64] |
Ito S, Chen C, Satoh J, et al. Dietary phytochemicals regulate whole-body CYP1A1 expression through an arylhydrocarbon receptor nuclear translocator-dependent system in gut[J]. J Clin Invest, 2007, 117(7): 1940-1950.
pmid: 17607366 |
| [65] |
Schipke J, Vital M, Schnapper-Isl A, et al. Spermidine and voluntary activity exert differential effects on sucrose- compared with fat-induced systemic changes in male mice[J]. J Nutr, 2019, 149(3): 451-462.
doi: 10.1093/jn/nxy272 pmid: 30715385 |
| [66] |
Carriche GM, Almeida L, Stüve P, et al. Regulating T-cell differentiation through the polyamine spermidine[J]. J Allergy Clin Immunol, 2021, 147(1): 335-348.e11.
doi: 10.1016/j.jaci.2020.04.037 pmid: 32407834 |
| [67] |
Wang J, Tan BE, Li JJ, et al. Regulatory role of l-proline in fetal pig growth and intestinal epithelial cell proliferation[J]. Anim Nutr, 2020, 6(4): 438-446.
doi: 10.1016/j.aninu.2020.07.001 pmid: 33364460 |
| [68] |
ter Steege JC, Buurman WA, Forget PP. Spermine induces maturation of the immature intestinal immune system in neonatal mice[J]. J Pediatr Gastroenterol Nutr, 1997, 25(3): 332-340.
doi: 10.1097/00005176-199709000-00017 URL |
| [69] |
Moinard C, Cynober L, de Bandt JP. Polyamines: metabolism and implications in human diseases[J]. Clin Nutr, 2005, 24(2): 184-197.
doi: 10.1016/j.clnu.2004.11.001 pmid: 15784477 |
| [70] |
Pérez-Cano FJ, González-Castro A, Castellote C, et al. Influence of breast milk polyamines on suckling rat immune system maturation[J]. Dev Comp Immunol, 2010, 34(2): 210-218.
doi: 10.1016/j.dci.2009.10.001 pmid: 19825390 |
| [71] |
Yoo JY, Groer M, Dutra SVO, et al. Gut microbiota and immune system interactions[J]. Microorganisms, 2020, 8(10): 1587.
doi: 10.3390/microorganisms8101587 URL |
| [1] | ZHOU Ai-ting, PENG Rui-qi, WANG Fang, WU Jian-rong, MA Huan-cheng. Analysis of Metabolic Differences of Biocontrol Strain DZY6715 at Different Growth Stages [J]. Biotechnology Bulletin, 2023, 39(9): 225-235. |
| [2] | XIONG Shu-qi. Towards the Understanding on the Physiological Functions of Bile Acids and Interactions with Gut Microbiota [J]. Biotechnology Bulletin, 2023, 39(4): 187-200. |
| [3] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [4] | HE Ya-lun, ZENG Li-rong, LIU Xiong, ZHANG Ling, WANG Qiong. Effects of High-dose Tannic Acid on the Intestinal Barrier Function and Gut Microbiota in Mice [J]. Biotechnology Bulletin, 2022, 38(4): 278-287. |
| [5] | YANG Yu-ping, ZHANG Xia, WANG Chong-chong, WANG Xiao-yan. Study on Urine Metabolomics in Rats of Different Ages [J]. Biotechnology Bulletin, 2022, 38(2): 166-172. |
| [6] | YANG Rui-xian, LIU Ping, WANG Zu-hua, RUAN Bao-shuo, WANG Zhi-da. Analysis of Antimicrobial Active Metabolites from Antagonistic Strains Against Fusarium solani [J]. Biotechnology Bulletin, 2022, 38(2): 57-66. |
| [7] | LIU Chuan-he, HE Han, HE Xiu-gu, LAI Qiu-qin, LIU Kai, SHAO Xue-hua, LAI Duo, KUANG Shi-zi, XIAO Wei-qiang. Unveiling the Mechanisms of Pineapple Responding to Anti-chilling by Gauze Covering in Winter via Transcriptome and Metabolome Profiling [J]. Biotechnology Bulletin, 2022, 38(11): 58-69. |
| [8] | LIU Chuan-he, HE Han, HE Xiu-gu, LIU Kai, SHAO Xue-hua, LAI Duo, KUANG Shi-zi, XIAO Wei-qiang. Analysis of Differential Metabolites and Bacterial Community Structure in Soils of a Pineapple Orchard in Different Continuous-cropping Years [J]. Biotechnology Bulletin, 2021, 37(8): 162-175. |
| [9] | WANG Nan, SU Yu, LIU Wen-jie, FENG Ming, MAO Yu, ZHANG Xin-guo. Research Progress on Active Compounds Against Drug-resistant Microorganism from Plant Endophytes [J]. Biotechnology Bulletin, 2021, 37(8): 263-274. |
| [10] | LIANG Zhen-ting, TANG Ting. Effects of Endophytes on Biosynthesis of Secondary Metabolites and Stress Tolerance in Plants [J]. Biotechnology Bulletin, 2021, 37(8): 35-45. |
| [11] | LI Hai-chao, XIE Fei, ZHANG Yuan-qi, GUAN Ruo-bing. Effects of Resistant and Sensitive Rice Varieties on Gut Microbiota of Nilaparvata lugens [J]. Biotechnology Bulletin, 2021, 37(3): 1-9. |
| [12] | HUANG Xiao-dan, CHEN Meng-yu, HUANG Wen-jie, ZHANG Ming-wei, YAN Shi-juan. Progress Based on Metabolomics:Plant Polyphenols and Their Gut Health Benefit [J]. Biotechnology Bulletin, 2021, 37(1): 123-136. |
| [13] | LIU Yu, DING Qian-wen, RAN Chao, YANG Ya-lin, WANG An-ran, ZHANG Hong-ling, ZHANG Jin-xiong, LI Jie, Rolf Erik OLSEN, Einar RINGØ, ZHANG Zhen, ZHOU Zhi-gang. Research Advances on Short-chain Fatty Acids of Metabolites of Gut Microbiota in Aquatic Animals [J]. Biotechnology Bulletin, 2020, 36(2): 58-64. |
| [14] | WU Qin, XU Zi-yang, LIU Li-ping, ZHANG Wen-ying, SONG Si-yuan. Role of Gut Microbiota in Stress-induced Hypertension in Rats [J]. Biotechnology Bulletin, 2020, 36(2): 83-90. |
| [15] | ZHAO Jiang-hua, FANG Huan, ZHANG Da-wei. Research Progress in Biosynthesis of Secondary Metabolites of Microorganisms [J]. Biotechnology Bulletin, 2020, 36(11): 141-147. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||