








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (10): 268-280.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0178
Previous Articles Next Articles
Received:2023-03-02
Online:2023-10-26
Published:2023-11-28
TANG Bi-yao FU Xue-peng. Sequencing Analysis of the Whole Genome of Streptomyces sp. FXP04[J]. Biotechnology Bulletin, 2023, 39(10): 268-280.
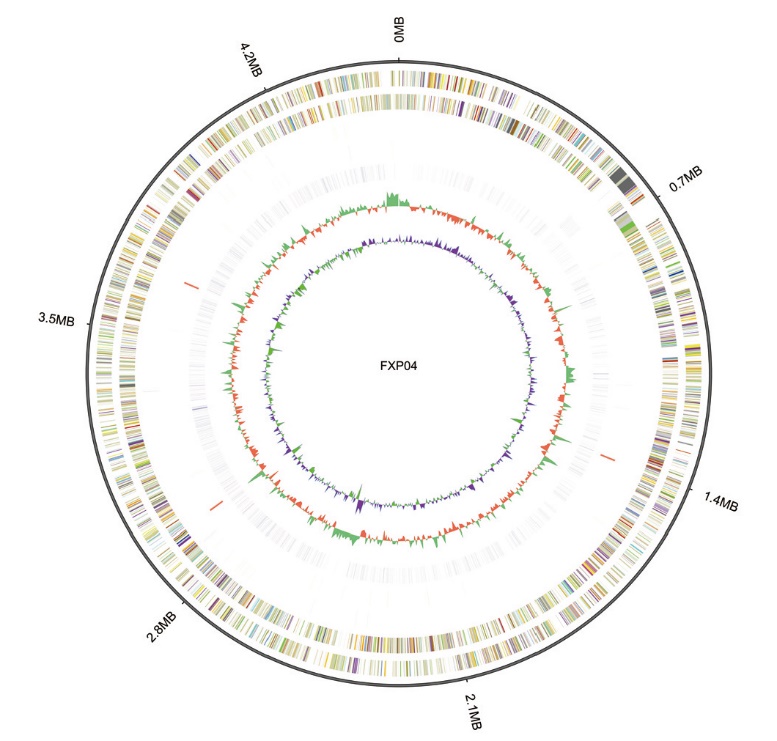
Fig. 1 Whole genome of strain FXP04 From outside to inside, it is: 1: the scale of genome(Mb); 2: positive chain gene; 3: negative chain gene; 4: positive chain long-chain non-coding RNA; 5: negative chain long-chain non-coding RNA; 6: repeat; 7: GC content; 8: GC offset
| Gene_ID | Align length/bp | Gene_ID | Align length/bp | Gene_ID | Align length/bp | Gene_ID | Align length/bp | |||
|---|---|---|---|---|---|---|---|---|---|---|
| FXP04GL000034 | 145 | FXP04GL001071 | 217 | FXP04GL002181 | 273 | FXP04GL003852 | 312 | |||
| FXP04GL000110 | 421 | FXP04GL001084 | 256 | FXP04GL002246 | 245 | FXP04GL003853 | 236 | |||
| FXP04GL000136 | 306 | FXP04GL001105 | 240 | FXP04GL002251 | 220 | FXP04GL003854 | 216 | |||
| FXP04GL000172 | 214 | FXP04GL001156 | 979 | FXP04GL002258 | 250 | FXP04GL003869 | 162 | |||
| FXP04GL000206 | 334 | FXP04GL001158 | 209 | FXP04GL002399 | 119 | FXP04GL003888 | 175 | |||
| FXP04GL000363 | 214 | FXP04GL001159 | 549 | FXP04GL002411 | 192 | FXP04GL003982 | 283 | |||
| FXP04GL000408 | 388 | FXP04GL001160 | 407 | FXP04GL002412 | 198 | FXP04GL004032 | 236 | |||
| FXP04GL000514 | 627 | FXP04GL001161 | 260 | FXP04GL002431 | 191 | FXP04GL004050 | 405 | |||
| FXP04GL000547 | 1999 | FXP04GL001163 | 331 | FXP04GL002686 | 60 | FXP04GL004060 | 268 | |||
| FXP04GL000548 | 1445 | FXP04GL001165 | 239 | FXP04GL002830 | 108 | FXP04GL004064 | 172 | |||
| FXP04GL000549 | 668 | FXP04GL001238 | 257 | FXP04GL002969 | 90 | FXP04GL004069 | 217 | |||
| FXP04GL000551 | 461 | FXP04GL001239 | 376 | FXP04GL002999 | 244 | FXP04GL004082 | 230 | |||
| FXP04GL000564 | 2121 | FXP04GL001240 | 342 | FXP04GL003031 | 402 | FXP04GL004108 | 134 | |||
| FXP04GL000565 | 1865 | FXP04GL001265 | 205 | FXP04GL003068 | 217 | FXP04GL004109 | 181 | |||
| FXP04GL000566 | 1515 | FXP04GL001280 | 241 | FXP04GL003104 | 222 | FXP04GL004113 | 288 | |||
| FXP04GL000569 | 565 | FXP04GL001338 | 430 | FXP04GL003123 | 89 | FXP04GL004121 | 106 | |||
| FXP04GL000605 | 240 | FXP04GL001411 | 550 | FXP04GL003154 | 149 | FXP04GL004165 | 244 | |||
| FXP04GL000609 | 708 | FXP04GL001419 | 145 | FXP04GL003179 | 228 | FXP04GL004178 | 80 | |||
| FXP04GL000639 | 403 | FXP04GL001559 | 517 | FXP04GL003222 | 426 | FXP04GL004456 | 135 | |||
| FXP04GL000646 | 217 | FXP04GL001670 | 300 | FXP04GL003249 | 287 | FXP04GL004473 | 237 | |||
| FXP04GL000652 | 234 | FXP04GL001676 | 232 | FXP04GL003255 | 338 | FXP04GL004474 | 240 | |||
| FXP04GL000653 | 501 | FXP04GL001677 | 307 | FXP04GL003405 | 240 | FXP04GL004485 | 94 | |||
| FXP04GL000663 | 306 | FXP04GL001711 | 222 | FXP04GL003459 | 185 | FXP04GL004486 | 135 | |||
| FXP04GL000684 | 110 | FXP04GL001715 | 368 | FXP04GL003460 | 134 | FXP04GL004571 | 128 | |||
| FXP04GL000722 | 73 | FXP04GL001782 | 476 | FXP04GL003505 | 226 | FXP04GL004641 | 415 | |||
| FXP04GL000785 | 212 | FXP04GL001792 | 135 | FXP04GL003545 | 98 | FXP04GL004644 | 70 | |||
| FXP04GL000792 | 334 | FXP04GL001793 | 300 | FXP04GL003552 | 92 | FXP04GL004796 | 110 | |||
| FXP04GL000926 | 200 | FXP04GL001812 | 100 | FXP04GL003574 | 213 | FXP04GL004808 | 237 | |||
| FXP04GL000927 | 443 | FXP04GL001849 | 274 | FXP04GL003661 | 171 | FXP04GL004815 | 141 | |||
| FXP04GL000969 | 219 | FXP04GL001901 | 260 | FXP04GL003662 | 273 | FXP04GL004866 | 250 | |||
| FXP04GL000972 | 249 | FXP04GL001938 | 129 | FXP04GL003663 | 259 | FXP04GL004870 | 90 | |||
| FXP04GL000973 | 503 | FXP04GL001947 | 163 | FXP04GL003664 | 282 | FXP04GL004871 | 103 | |||
| FXP04GL001037 | 306 | FXP04GL001972 | 130 | FXP04GL003701 | 41 | FXP04GL004959 | 324 | |||
| FXP04GL001053 | 162 | FXP04GL002050 | 218 | FXP04GL003794 | 393 | |||||
| FXP04GL001062 | 237 | FXP04GL002068 | 268 | FXP04GL003807 | 166 |
Table 1 Annotation results of strain FXP04 in VFDB database
| Gene_ID | Align length/bp | Gene_ID | Align length/bp | Gene_ID | Align length/bp | Gene_ID | Align length/bp | |||
|---|---|---|---|---|---|---|---|---|---|---|
| FXP04GL000034 | 145 | FXP04GL001071 | 217 | FXP04GL002181 | 273 | FXP04GL003852 | 312 | |||
| FXP04GL000110 | 421 | FXP04GL001084 | 256 | FXP04GL002246 | 245 | FXP04GL003853 | 236 | |||
| FXP04GL000136 | 306 | FXP04GL001105 | 240 | FXP04GL002251 | 220 | FXP04GL003854 | 216 | |||
| FXP04GL000172 | 214 | FXP04GL001156 | 979 | FXP04GL002258 | 250 | FXP04GL003869 | 162 | |||
| FXP04GL000206 | 334 | FXP04GL001158 | 209 | FXP04GL002399 | 119 | FXP04GL003888 | 175 | |||
| FXP04GL000363 | 214 | FXP04GL001159 | 549 | FXP04GL002411 | 192 | FXP04GL003982 | 283 | |||
| FXP04GL000408 | 388 | FXP04GL001160 | 407 | FXP04GL002412 | 198 | FXP04GL004032 | 236 | |||
| FXP04GL000514 | 627 | FXP04GL001161 | 260 | FXP04GL002431 | 191 | FXP04GL004050 | 405 | |||
| FXP04GL000547 | 1999 | FXP04GL001163 | 331 | FXP04GL002686 | 60 | FXP04GL004060 | 268 | |||
| FXP04GL000548 | 1445 | FXP04GL001165 | 239 | FXP04GL002830 | 108 | FXP04GL004064 | 172 | |||
| FXP04GL000549 | 668 | FXP04GL001238 | 257 | FXP04GL002969 | 90 | FXP04GL004069 | 217 | |||
| FXP04GL000551 | 461 | FXP04GL001239 | 376 | FXP04GL002999 | 244 | FXP04GL004082 | 230 | |||
| FXP04GL000564 | 2121 | FXP04GL001240 | 342 | FXP04GL003031 | 402 | FXP04GL004108 | 134 | |||
| FXP04GL000565 | 1865 | FXP04GL001265 | 205 | FXP04GL003068 | 217 | FXP04GL004109 | 181 | |||
| FXP04GL000566 | 1515 | FXP04GL001280 | 241 | FXP04GL003104 | 222 | FXP04GL004113 | 288 | |||
| FXP04GL000569 | 565 | FXP04GL001338 | 430 | FXP04GL003123 | 89 | FXP04GL004121 | 106 | |||
| FXP04GL000605 | 240 | FXP04GL001411 | 550 | FXP04GL003154 | 149 | FXP04GL004165 | 244 | |||
| FXP04GL000609 | 708 | FXP04GL001419 | 145 | FXP04GL003179 | 228 | FXP04GL004178 | 80 | |||
| FXP04GL000639 | 403 | FXP04GL001559 | 517 | FXP04GL003222 | 426 | FXP04GL004456 | 135 | |||
| FXP04GL000646 | 217 | FXP04GL001670 | 300 | FXP04GL003249 | 287 | FXP04GL004473 | 237 | |||
| FXP04GL000652 | 234 | FXP04GL001676 | 232 | FXP04GL003255 | 338 | FXP04GL004474 | 240 | |||
| FXP04GL000653 | 501 | FXP04GL001677 | 307 | FXP04GL003405 | 240 | FXP04GL004485 | 94 | |||
| FXP04GL000663 | 306 | FXP04GL001711 | 222 | FXP04GL003459 | 185 | FXP04GL004486 | 135 | |||
| FXP04GL000684 | 110 | FXP04GL001715 | 368 | FXP04GL003460 | 134 | FXP04GL004571 | 128 | |||
| FXP04GL000722 | 73 | FXP04GL001782 | 476 | FXP04GL003505 | 226 | FXP04GL004641 | 415 | |||
| FXP04GL000785 | 212 | FXP04GL001792 | 135 | FXP04GL003545 | 98 | FXP04GL004644 | 70 | |||
| FXP04GL000792 | 334 | FXP04GL001793 | 300 | FXP04GL003552 | 92 | FXP04GL004796 | 110 | |||
| FXP04GL000926 | 200 | FXP04GL001812 | 100 | FXP04GL003574 | 213 | FXP04GL004808 | 237 | |||
| FXP04GL000927 | 443 | FXP04GL001849 | 274 | FXP04GL003661 | 171 | FXP04GL004815 | 141 | |||
| FXP04GL000969 | 219 | FXP04GL001901 | 260 | FXP04GL003662 | 273 | FXP04GL004866 | 250 | |||
| FXP04GL000972 | 249 | FXP04GL001938 | 129 | FXP04GL003663 | 259 | FXP04GL004870 | 90 | |||
| FXP04GL000973 | 503 | FXP04GL001947 | 163 | FXP04GL003664 | 282 | FXP04GL004871 | 103 | |||
| FXP04GL001037 | 306 | FXP04GL001972 | 130 | FXP04GL003701 | 41 | FXP04GL004959 | 324 | |||
| FXP04GL001053 | 162 | FXP04GL002050 | 218 | FXP04GL003794 | 393 | |||||
| FXP04GL001062 | 237 | FXP04GL002068 | 268 | FXP04GL003807 | 166 |
| Gene_ID | Subject ID | Resistance type | Resistance requirement | Antibiotic resistance |
|---|---|---|---|---|
| FXP04GL000228 | ardb_195 | pur8 | -- | Puromycin |
| FXP04GL000655 | ardb_2322 | bcra | bcrc | Bacitracin |
| FXP04GL001363 | ardb_1271 | emre | emrd | Aminoglycoside |
| FXP04GL001966 | ardb_195 | pur8 | -- | Puromycin |
| FXP04GL002045 | ardb_2828 | tcma | -- | Tetracenomycin_c |
| FXP04GL002950 | ardb_640 | ykkd | ykkc | Na_antimicrobials |
| FXP04GL003556 | ardb_1951 | cml_e6 | -- | Chloramphenicol |
| FXP04GL003691 | ardb_1951 | cml_e6 | -- | Chloramphenicol |
| FXP04GL004392 | ardb_2612 | aph6ib | -- | Streptomycin |
| FXP04GL004405 | ardb_195 | pur8 | -- | Puromycin |
| FXP04GL004638 | ardb_195 | pur8 | -- | Puromycin |
| FXP04GL004787 | ardb_1043 | vanha | vana,vanxa,vanya, vanra,vansa | Vancomycin, Teicoplanin |
| FXP04GL004788 | ardb_2982 | vand | vanhd,vanyd,vanxd,vanrd,vansd | Vancomycin, Teicoplanin |
| FXP04GL004789 | ardb_993 | vanxd | vand,vanhd,vanyd,vanrd,vansd | Vancomycin, Teicoplanin |
Table 2 Annotation results of strain FXP04 in ARDB database
| Gene_ID | Subject ID | Resistance type | Resistance requirement | Antibiotic resistance |
|---|---|---|---|---|
| FXP04GL000228 | ardb_195 | pur8 | -- | Puromycin |
| FXP04GL000655 | ardb_2322 | bcra | bcrc | Bacitracin |
| FXP04GL001363 | ardb_1271 | emre | emrd | Aminoglycoside |
| FXP04GL001966 | ardb_195 | pur8 | -- | Puromycin |
| FXP04GL002045 | ardb_2828 | tcma | -- | Tetracenomycin_c |
| FXP04GL002950 | ardb_640 | ykkd | ykkc | Na_antimicrobials |
| FXP04GL003556 | ardb_1951 | cml_e6 | -- | Chloramphenicol |
| FXP04GL003691 | ardb_1951 | cml_e6 | -- | Chloramphenicol |
| FXP04GL004392 | ardb_2612 | aph6ib | -- | Streptomycin |
| FXP04GL004405 | ardb_195 | pur8 | -- | Puromycin |
| FXP04GL004638 | ardb_195 | pur8 | -- | Puromycin |
| FXP04GL004787 | ardb_1043 | vanha | vana,vanxa,vanya, vanra,vansa | Vancomycin, Teicoplanin |
| FXP04GL004788 | ardb_2982 | vand | vanhd,vanyd,vanxd,vanrd,vansd | Vancomycin, Teicoplanin |
| FXP04GL004789 | ardb_993 | vanxd | vand,vanhd,vanyd,vanrd,vansd | Vancomycin, Teicoplanin |
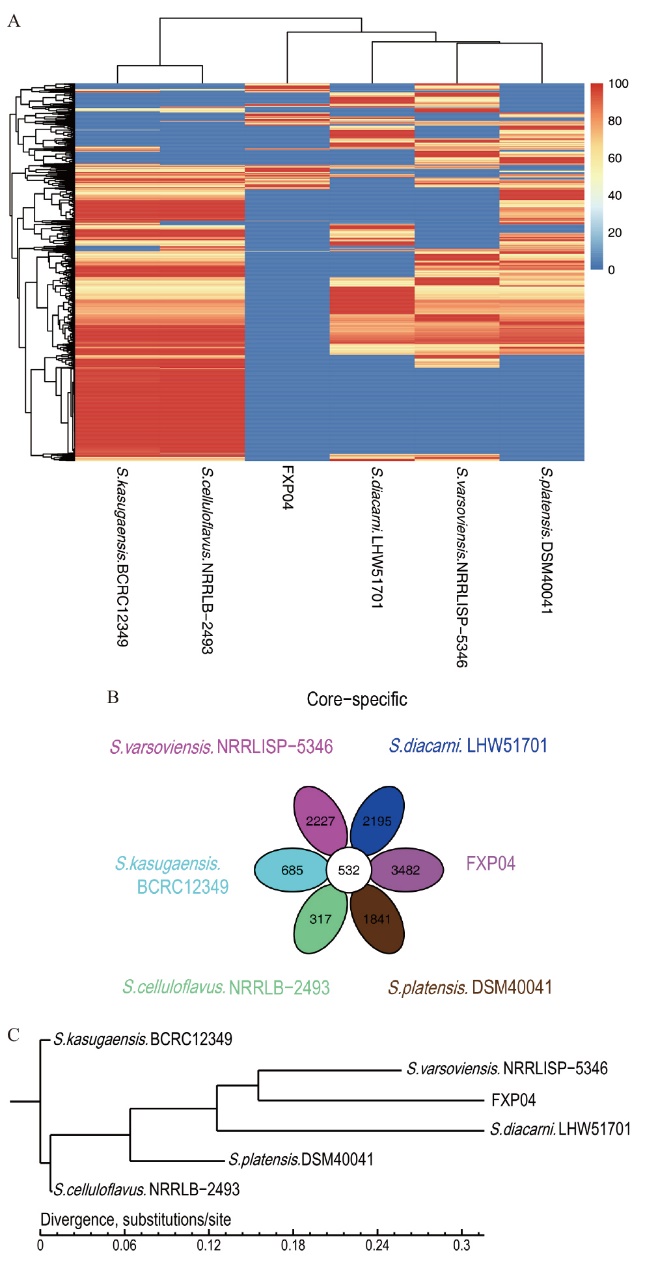
Fig. 5 Genome Core/Pan analysis results among 6 strains A: The heat map of the homologous relationship of Dispensable Gene. B: Venn diagram of Pan homology. C: Phylogenetic tree based on Core Gene homology relationship
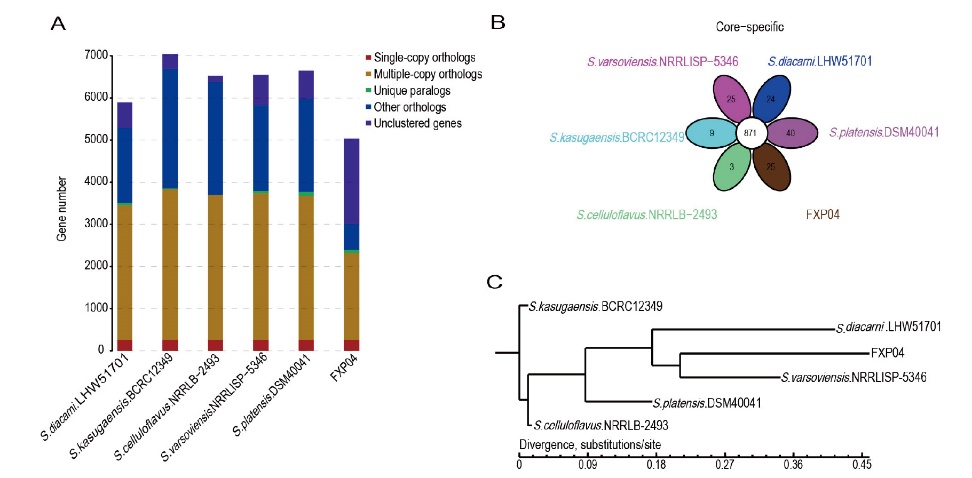
Fig. 6 Genome Gene family analysis results among 6 strains A: Homologous gene clustering. B: Venn diagram of Gene family homology. C: Phylogenetic tree based on homologous relationship of Gene Family

Fig. 7 Collinearity analysis between FXP04 and S.varsoviensis NRRLISP-5346 A: Amino acid collinearity diagram. B: Linear collinearity diagram of nucleic acid
| 簇编号 Cluster ID | 基因起始-终止位置 Start-stop position of gene | 基因数目 Number of genes | 簇类型 Cluster type | 已知基因簇 Known cluster | 相似度 Similarity/% | 来源 Resources |
|---|---|---|---|---|---|---|
| Cluster 1 | 94744-113290 | 15 | Terpene | Petrichorin A /Petrichorin B | 24 | Lentzea flaviverrucosa |
| Cluster 2 | 293232-340393 | 27 | NRPS | - | - | - |
| Cluster 3 | 570305-665318 | 54 | T1PKS | Piericidin A1 | 100 | Candidatus Streptomyces philanthi bv. triangulum |
| Cluster 4 | 666624-764035 | 47 | T1PKS, transAT-PKS, PKS-like | Psymberin /Irciniastatin B | 31 | Uncultured bacterium psy1 |
| Cluster 5 | 897552-922003 | 23 | Terpene | Hopene | 61 | Streptomyces coelicolor A3(2) |
| Cluster 6 | 1024430-1065437 | 31 | NRPS, NAPAA | Stenothricin | 13 | Streptomyces filamentosus NRRL 15998 |
| Cluster 7 | 1268236-1317622 | 37 | NRPS, NRP-metallophore | Griseobactin | 53 | Streptomyces sp. ATCC 700974 |
| Cluster 8 | 1475651-1486055 | 9 | Ectoine | Ectoine | 100 | Streptomyces sp. strain ID38640 |
| Cluster 9 | 1945067-1985405 | 46 | T3PKS | s56-p1 | 11 | Streptomyces sp. SoC090715LN-17 |
| Cluster 10 | 2502623-2524096 | 24 | Lassopeptide | - | - | - |
| Cluster 11 | 2586833-2624623 | 45 | Arylpolyene | Aurachin C/Aurachin D/Aurachin SS | 13 | Streptomyces sp. |
| Cluster 12 | 3558109-3579044 | 29 | - | - | - | - |
| Cluster 13 | 3811714-3852580 | 49 | Arylpolyene | Youssoufene A1/B1/B2/B3/B4 | 88 | Streptomyces youssoufiensis |
Table 3 Identification results of synthetic regions of secondary metabolites
| 簇编号 Cluster ID | 基因起始-终止位置 Start-stop position of gene | 基因数目 Number of genes | 簇类型 Cluster type | 已知基因簇 Known cluster | 相似度 Similarity/% | 来源 Resources |
|---|---|---|---|---|---|---|
| Cluster 1 | 94744-113290 | 15 | Terpene | Petrichorin A /Petrichorin B | 24 | Lentzea flaviverrucosa |
| Cluster 2 | 293232-340393 | 27 | NRPS | - | - | - |
| Cluster 3 | 570305-665318 | 54 | T1PKS | Piericidin A1 | 100 | Candidatus Streptomyces philanthi bv. triangulum |
| Cluster 4 | 666624-764035 | 47 | T1PKS, transAT-PKS, PKS-like | Psymberin /Irciniastatin B | 31 | Uncultured bacterium psy1 |
| Cluster 5 | 897552-922003 | 23 | Terpene | Hopene | 61 | Streptomyces coelicolor A3(2) |
| Cluster 6 | 1024430-1065437 | 31 | NRPS, NAPAA | Stenothricin | 13 | Streptomyces filamentosus NRRL 15998 |
| Cluster 7 | 1268236-1317622 | 37 | NRPS, NRP-metallophore | Griseobactin | 53 | Streptomyces sp. ATCC 700974 |
| Cluster 8 | 1475651-1486055 | 9 | Ectoine | Ectoine | 100 | Streptomyces sp. strain ID38640 |
| Cluster 9 | 1945067-1985405 | 46 | T3PKS | s56-p1 | 11 | Streptomyces sp. SoC090715LN-17 |
| Cluster 10 | 2502623-2524096 | 24 | Lassopeptide | - | - | - |
| Cluster 11 | 2586833-2624623 | 45 | Arylpolyene | Aurachin C/Aurachin D/Aurachin SS | 13 | Streptomyces sp. |
| Cluster 12 | 3558109-3579044 | 29 | - | - | - | - |
| Cluster 13 | 3811714-3852580 | 49 | Arylpolyene | Youssoufene A1/B1/B2/B3/B4 | 88 | Streptomyces youssoufiensis |
| [1] |
Procópio RE, Silva IR, Martins MK, et al. Antibiotics produced by Streptomyces[J]. Braz J Infect Dis, 2012, 16(5): 466-471.
doi: 10.1016/j.bjid.2012.08.014 pmid: 22975171 |
| [2] |
Dharmaraj S. Marine Streptomyces as a novel source of bioactive substances[J]. World J Microbiol Biotechnol, 2010, 26(12): 2123-2139.
doi: 10.1007/s11274-010-0415-6 URL |
| [3] |
Arasu MV, Duraipandiyan V, Ignacimuthu S. Antibacterial and antifungal activities of polyketide metabolite from marine Streptomyces sp. AP-123 and its cytotoxic effect[J]. Chemosphere, 2013, 90(2): 479-487.
doi: 10.1016/j.chemosphere.2012.08.006 URL |
| [4] |
Kim JD, Han JW, Lee SC, et al. Disease control effect of strevertenes produced by Streptomyces psammoticus against tomato Fusarium wilt[J]. J Agric Food Chem, 2011, 59(5): 1893-1899.
doi: 10.1021/jf1038585 URL |
| [5] |
胡磊, 牛世全, 景彩虹, 等. 拮抗油菜菌核病菌的链霉菌分离筛选与鉴定[J]. 中国油料作物学报, 2013, 35(1): 69-73.
doi: 10.7505/j.issn.1007-9084.2013.01.012 |
| Hu L, Niu SQ, Jing CH, et al. Identification of Streptomyces antagonizing oilseed rape pathogen Sclerotinia sclerotiorum[J]. Chin J Oil Crop Sci, 2013, 35(1): 69-73. | |
| [6] |
Fu XP, Liu S, Ru JR, et al. Biological control of potato late blight by Streptomyces sp. FXP04 and potential role of secondary metabolites[J]. Biol Contr, 2022, 169: 104891.
doi: 10.1016/j.biocontrol.2022.104891 URL |
| [7] | 付学鹏, 沈童飞, 等. 链霉菌株Streptomyces sp.FXP04对水稻种子萌发和幼苗生长的影响[J]. 作物杂志, 2020(6): 163-169. |
| Fu XP, Shen TF, et al. Effects of Streptomyces sp. FXP04 on seed germination and seedling growth of rice[J]. Crops, 2020(6): 163-169. | |
| [8] | 丁秀芹. 微生物基因组研究进展[J]. 科技资讯, 2009, 7(5): 11-12. |
| Ding XQ. Research progress of microbial genome[J]. Sci Technol Inf, 2009, 7(5): 11-12. | |
| [9] | 高圣风, 徐毕爽, 等. 生防菌株Bacillus velezensis Z全基因组测序分析[J]. 热带作物学报, 2021, 42(5): 1216-1222. |
| Gao SF, Xu BS, et al. Whole genome sequencing and analysis of the bio-control strain Bacillus velezensis Z[J]. Chin J Trop Crops, 2021, 42(5): 1216-1222. | |
| [10] |
Fu LM, Niu BF, Zhu ZW, et al. CD-HIT: accelerated for clustering the next-generation sequencing data[J]. Bioinformatics, 2012, 28(23): 3150-3152.
doi: 10.1093/bioinformatics/bts565 pmid: 23060610 |
| [11] |
Nandi T, Ong C, et al. A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence[J]. PLoS Pathog, 2010, 6(4): e1000845.
doi: 10.1371/journal.ppat.1000845 URL |
| [12] |
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput[J]. Nucleic Acids Res, 2004, 32(5): 1792-1797.
doi: 10.1093/nar/gkh340 pmid: 15034147 |
| [13] |
Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity[J]. BMC Bioinformatics, 2004, 5: 113.
pmid: 15318951 |
| [14] |
Blin K, Wolf T, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification[J]. Nucleic Acids Res, 2017, 45(W1): W36-W41.
doi: 10.1093/nar/gkx319 URL |
| [15] |
Lagesen K, Hallin P, Rødland EA, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes[J]. Nucleic Acids Res, 2007, 35(9): 3100-3108.
doi: 10.1093/nar/gkm160 pmid: 17452365 |
| [16] |
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence[J]. Nucleic Acids Res, 1997, 25(5): 955-964.
doi: 10.1093/nar/25.5.955 pmid: 9023104 |
| [17] |
Benson G. Tandem repeats finder: a program to analyze DNA sequences[J]. Nucleic Acids Res, 1999, 27(2): 573-580.
doi: 10.1093/nar/27.2.573 pmid: 9862982 |
| [18] |
Zhou Y, Liang YJ, Lynch KH, et al. PHAST: a fast phage search tool[J]. Nucleic Acids Res, 2011, 39(Web Server issue): W347-W352.
doi: 10.1093/nar/gkr485 URL |
| [19] |
Luo YG, Lin SJ, et al. Regiospecific O-methylation of naphthoic acids catalyzed by NcsB1, an O-methyltransferase involved in the biosynthesis of the enediyne antitumor antibiotic neocarzinostatin[J]. J Biol Chem, 2008, 283(21): 14694-14702.
doi: 10.1074/jbc.M802206200 pmid: 18387946 |
| [20] | 尹明明, 陈代杰, 等. Eremomycin的发酵条件优化及发酵产物的分离纯化[J]. 微生物学杂志, 2010, 30(4): 22-26. |
| Yin MM, Chen DJ, et al. Fermentation conditions optimization, isolation and purification of eremomycin[J]. J Microbiol, 2010, 30(4): 22-26. | |
| [21] | 李永辉, 刘云, 王世春, 等. 大肠杆菌ppsA和tktA基因的串联表达[J]. 生物工程学报, 2003, 19(3): 301-306. |
| Li YH, Liu Y, Wang SC, et al. Co-expressions of phosphoenolpyruvate synthetase a(ppsA)and transketolase A(tktA)genes of Escherichia coli[J]. Chin J Biotechnol, 2003, 19(3): 301-306. | |
| [22] |
Liu Z, Zhang YN, Sun JN, et al. A novel soluble squalene-hopene cyclase and its application in efficient synthesis of hopene[J]. Front Bioeng Biotechnol, 2020, 8: 426.
doi: 10.3389/fbioe.2020.00426 URL |
| [23] | 黄天培, 潘洁茹, 张鸿声, 等. 扁座壳孢Jos009胞外代谢产物15α, 22-二羟何帕烷的抗菌活性[J]. 应用与环境生物学报, 2013, 19(5): 878-880. |
|
Huang TP, Pan JR, Zhang HS, et al. Antimicrobial activity of 15α, 22-dihydroxyhopane produced by Aschersonia placenta Jos009[J]. Chin J Appl Environ Biol, 2013, 19(5): 878-880.
doi: 10.3724/SP.J.1145.2013.00878 URL |
|
| [24] |
Chen Z, Washio T, Sato M, et al. Cytotoxic effects of several hopanoids on mouse leukemia L1210 and P388 cells[J]. Biol Pharm Bull, 1995, 18(3): 421-423.
pmid: 7550095 |
| [25] |
Dayanidhi DL, Somarelli JA, et al. Psymberin, a marine-derived natural product, induces cancer cell growth arrest and protein translation inhibition[J]. Front Med, 2022, 9: 999004.
doi: 10.3389/fmed.2022.999004 URL |
| [26] |
Li CS, Hu YF, Wu XH, et al. Discovery of unusual dimeric piperazyl cyclopeptides encoded by a Lentzea flaviverrucosa DSM 44664 biosynthetic supercluster[J]. Proc Natl Acad Sci USA, 2022, 119(17): e2117941119.
doi: 10.1073/pnas.2117941119 URL |
| [27] |
Patzer SI, Braun V. Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974[J]. J Bacteriol, 2010, 192(2): 426-435.
doi: 10.1128/JB.01250-09 pmid: 19915026 |
| [28] |
Matsuda K, Tomita T, Shin-Ya K, et al. Discovery of unprecedented hydrazine-forming machinery in bacteria[J]. J Am Chem Soc, 2018, 140(29): 9083-9086.
doi: 10.1021/jacs.8b05354 pmid: 30001119 |
| [29] |
Kunze B, Höfle G, Reichenbach H. The aurachins, new quinoline antibiotics from myxobacteria: production, physico-chemical and biological properties[J]. J Antibiot, 1987, 40(3): 258-265.
pmid: 3106289 |
| [30] |
Zhang M, Yang CL, et al. Aurachin SS, a new antibiotic from Streptomyces sp. NA04227[J]. J Antibiot, 2017, 70(7): 853-855.
doi: 10.1038/ja.2017.50 pmid: 28420868 |
| [31] |
Pastor JM, Salvador M, Argandoña M, et al. Ectoines in cell stress protection: uses and biotechnological production[J]. Biotechnol Adv, 2010, 28(6): 782-801.
doi: 10.1016/j.biotechadv.2010.06.005 pmid: 20600783 |
| [32] |
Shi J, Shi Y, Li JC, et al. In vitro reconstitution of cinnamoyl moiety reveals two distinct cyclases for benzene ring formation[J]. J Am Chem Soc, 2022, 144(17): 7939-7948.
doi: 10.1021/jacs.2c02855 URL |
| [33] |
Hall C, Wu M, Crane FL, et al. Piericidin A: a new inhibitor of mitochondrial electron transport[J]. Biochem Biophys Res Commun, 1966, 25(4): 373-377.
doi: 10.1016/0006-291X(66)90214-2 URL |
| [34] |
Zhou XF, Fenical W. The unique chemistry and biology of the piericidins[J]. J Antibiot, 2016, 69(8): 582-593.
doi: 10.1038/ja.2016.71 pmid: 27301663 |
| [35] |
Darrouzet E, Issartel JP, Lunardi J, et al. The 49-kDa subunit of NADH-ubiquinone oxidoreductase(Complex I)is involved in the binding of piericidin and rotenone, two quinone-related inhibitors[J]. FEBS Lett, 1998, 431(1): 34-38.
pmid: 9684860 |
| [36] | Li WL, Zhang WY, Cheng YJ, et al. Investigation of carbonyl amidation and O-methylation during biosynthesis of the pharmacophore pyridyl of antitumor piericidins[J]. Synth Syst Biotechnol, 2022, 7(3): 880-886. |
| [37] |
Zhou XF, Liang Z, Li KL, et al. Exploring the natural piericidins as anti-renal cell carcinoma agents targeting peroxiredoxin 1[J]. J Med Chem, 2019, 62(15): 7058-7069.
doi: 10.1021/acs.jmedchem.9b00598 pmid: 31298537 |
| [38] |
Azad SM, Jin Y, Ser HL, et al. Biological insights into the piericidin family of microbial metabolites[J]. J Appl Microbiol, 2022, 132(2): 772-784.
doi: 10.1111/jam.v132.2 URL |
| [39] | 何亚文, Mazhari A., 崔莹, 等. 高产杀粉蝶菌素的链霉菌鉴定及其拮抗水稻白叶枯菌活性研究[J]. 微生物学报, 2023, 63(4): 1447-1459. |
| He YW, Mazhari A, Cui Y, et al. A piericidin-producing Streptomyces strain: identification and antagonistic activity on Xanthomonas oryzae[J]. Acta Microbiol Sin, 2023, 63(4): 1447-1459. | |
| [40] | 卜庆廷, 毛旭明, 等. 链霉菌较小基因组研究进展及展望[J]. 生命科学, 2019, 31(4): 391-401. |
| Bu QT, Mao XM, et al. Progress and prospect of genome-minimized Streptomyces hosts[J]. Chin Bull Life Sci, 2019, 31(4): 391-401. |
| [1] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [2] | LI Xue-qi, ZHANG Su-jie, YU Man, HUANG Jin-guang, ZHOU Huan-bin. Establishment of CRISPR/CasX-based Genome Editing Technology in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 40-48. |
| [3] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [4] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [5] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [6] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| [7] | SHI Jia-xin, LIU Kai, ZHU Jin-jie, QI Xian-tao, XIE Chuan-xiao, LIU Chang-lin. Gene Editing Reshaping Maize Plant Type for Increasing Hybrid Yield [J]. Biotechnology Bulletin, 2023, 39(8): 62-69. |
| [8] | DU Dong-dong, QIAN Jing, LI Si-qi, LIU Wen-fei, WEI Xiang-li, LIU Chang-yong, LUO Rui-feng, KANG Li-chao. Whole Genome Sequencing and Analysis of Listeria monocytogenes Strain LMXJ15 [J]. Biotechnology Bulletin, 2023, 39(7): 298-306. |
| [9] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [10] | YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan [J]. Biotechnology Bulletin, 2023, 39(6): 233-247. |
| [11] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [12] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [13] | ZHOU Xiao-jie, YANG Si-qi, ZHANG Yi-wen, XU Jia-qi, YANG Sheng. CRISPR-associated Transposases and Their Applications in Bacterial Genome Editing [J]. Biotechnology Bulletin, 2023, 39(4): 49-58. |
| [14] | XIAO Xiao-jun, CHEN Ming, HAN De-peng, YU Pao-lan, ZHENG Wei, XIAO Guo-bin, ZHOU Qing-hong, ZHOU Hui-wen. Genome Wide Association Analysis of Thousand Seed Weight in Brassica napus L. [J]. Biotechnology Bulletin, 2023, 39(3): 143-151. |
| [15] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
