








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (10): 281-291.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0307
Previous Articles Next Articles
YANG Jun-zhao( ), ZHANG Xin-rui, SUN Qing-yang, ZHENG Fei(
), ZHANG Xin-rui, SUN Qing-yang, ZHENG Fei( )
)
Received:2023-04-06
Online:2023-10-26
Published:2023-11-28
Contact:
ZHENG Fei
E-mail:YJZbio@bjfu.edu.cn;zhengfei0718@bjfu.edu.cn
YANG Jun-zhao, ZHANG Xin-rui, SUN Qing-yang, ZHENG Fei. Affecting Mechanism of Loop B3 on the Function of GH7 Endoglucanase[J]. Biotechnology Bulletin, 2023, 39(10): 281-291.
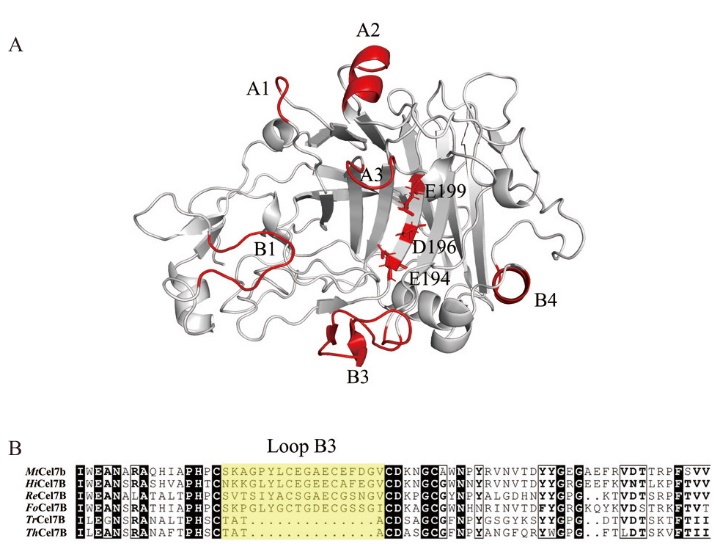
Fig. 1 Structural simulation(A)and amino acid sequence alignment(B)of MtCel7b A: Modeled structure of MtCel7b, loop structure in red, E194, D196 and E199 are catalytic triplets. B: The Loop region are marked by a yellow box. The strain source and PDB ID of the alignment sequence are HiCel7B(H. insolens, 6YOZ); ReCel7B(R. emersonii CBS 394.64, 6SU8); FoCel7B(F. oxysporum, 1OVW); TrCel7B(T. reesei, 1EG1); ThCel7B(T. harzianum CBS 226.95, 5W0A)
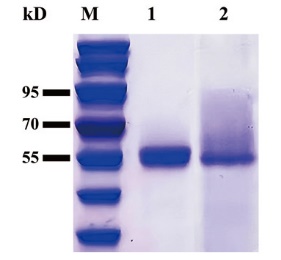
Fig. 2 SDS-PAGE analysis of wild-type MtCel7b and mutant B3cut M: Protein molecular weight standard; 1: purified protein of wild-type MtCel7b; 2: purified protein of mutant B3cut
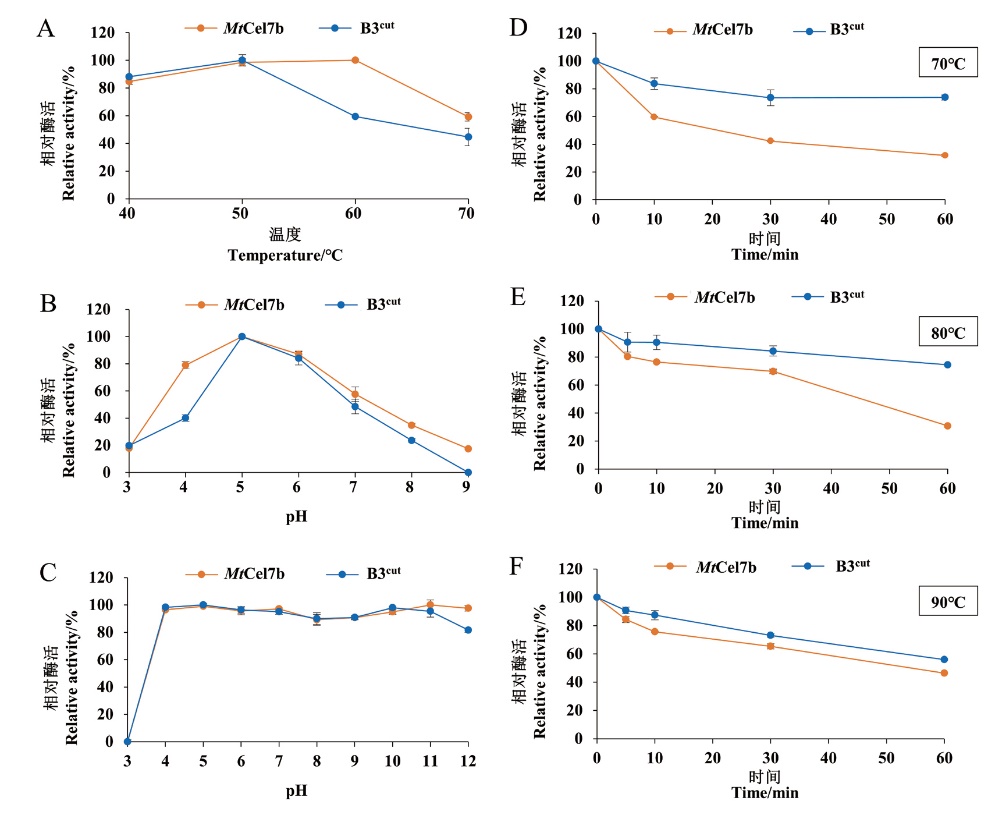
Fig. 3 Enzymatic properties of wild-type MtCel7b and mutant B3cut A: Optimal temperature-activity profile. B: Optimal pH. C: pH stability. D: Thermostability at 70℃. E: Thermostability at 80℃. F: Thermostability at 90℃
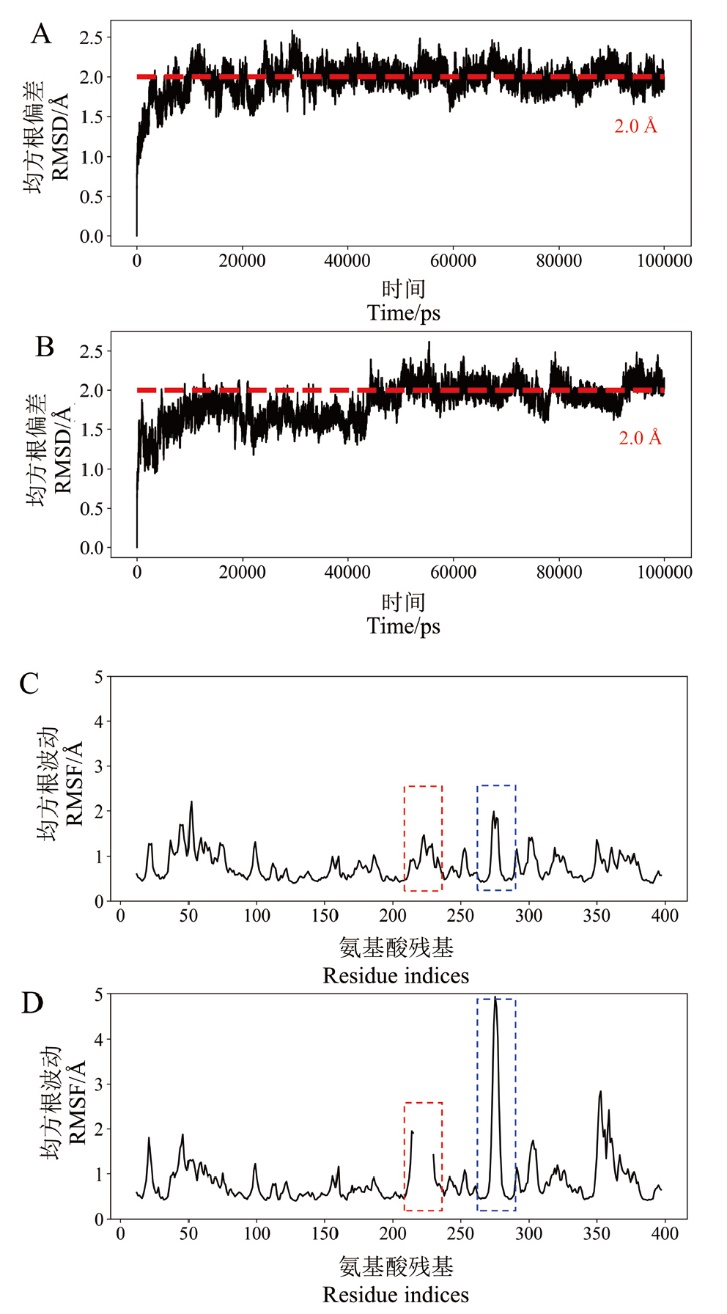
Fig. 6 RMSD and RMSF of wild-type MtCel7b and mutant B3cut A: RMSD of wild-type MtCel7b; B: RMSD of mutant B3cut; C: RMSF of wild-type MtCel7b; D: RMSF of mutant B3cut

Fig. 7 Trajectory principal component analysis plots and average structure of molecular dynamics simulations of wild-type MtCel7b and mutant B3cut A and B: Principal component analysis of the molecular dynamic simulation trajectory of wild-type MtCel7b and mutant B3cut. C and D: Protein surface of wild-type MtCel7b and mutant B3cut. E and F: Amino acid residues around the catalytic cleft of wild-type MtCel7b and mutant B3cut
| [1] |
Chen Y, Huang JW, Chen CC, et al. Crystallization and preliminary X-ray diffraction analysis of an endo-1,4-β-D-glucanase from Aspergillus aculeatus F-50[J]. Acta Crystallogr F Struct Biol Commun, 2015, 71(Pt 4): 397-400.
doi: 10.1107/S2053230X15003659 URL |
| [2] |
Bhat MK, Bhat S. Cellulose degrading enzymes and their potential industrial applications[J]. Biotechnol Adv, 1997, 15(3/4): 583-620.
doi: 10.1016/S0734-9750(97)00006-2 URL |
| [3] |
Kitamoto N, Go M, Shibayama T, et al. Molecular cloning, purification and characterization of two endo-1, 4-beta-glucanases from Aspergillus oryzae KBN616[J]. Appl Microbiol Biotechnol, 1996, 46(5/6): 538-544.
doi: 10.1007/s002530050857 URL |
| [4] |
Takashima S, Nakamura A, Hidaka M, et al. Cloning, sequencing, and expression of the cellulase genes of Humicola grisea var. thermoidea[J]. J Biotechnol, 1996, 50(2/3): 137-147.
doi: 10.1016/0168-1656(96)01555-6 URL |
| [5] |
Luo HY, Yang J, Yang PL, et al. Gene cloning and expression of a new acidic family 7 endo-beta-1, 3-1, 4-glucanase from the acidophilic fungus Bispora sp. MEY-1[J]. Appl Microbiol Biotechnol, 2010, 85(4): 1015-1023.
doi: 10.1007/s00253-009-2119-0 URL |
| [6] |
Bernardi AV, de Gouvêa PF, Gerolamo LE, et al. Functional characterization of GH7 endo-1,4-β-glucanase from Aspergillus fumigatus and its potential industrial application[J]. Protein Expr Purif, 2018, 150: 1-11.
doi: 10.1016/j.pep.2018.04.016 URL |
| [7] |
Kleywegt GJ, Zou JY, Divne C, et al. The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 Å resolution, and a comparison with related enzymes[J]. J Mol Biol, 1997, 272(3): 383-397.
pmid: 9325098 |
| [8] |
Sulzenbacher G, Driguez H, Henrissat B, et al. Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: substrate distortion gives rise to the preferred axial orientation for the leaving group[J]. Biochemistry, 1996, 35(48): 15280-15287.
pmid: 8952478 |
| [9] |
MacKenzie LF, Sulzenbacher G, Divne C, et al. Crystal structure of the family 7 endoglucanase I(Cel7B)from Humicola insolens at 2.2 Å resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate[J]. Biochem J, 1998, 335(Pt 2)(Pt 2):409-416.
doi: 10.1042/bj3350409 URL |
| [10] |
Sonoda MT, Godoy AS, Pellegrini VOA, et al. Structure and dynamics of Trichoderma harzianum Cel7B suggest molecular architecture adaptations required for a wide spectrum of activities on plant cell wall polysaccharides[J]. Biochim Biophys Acta Gen Subj, 2019, 1863(6): 1015-1026.
doi: 10.1016/j.bbagen.2019.03.013 URL |
| [11] |
Schiano-di-Cola C, Kołaczkowski B, Sørensen TH, et al. Structural and biochemical characterization of a family 7 highly thermostable endoglucanase from the fungus Rasamsonia emersonii[J]. FEBS J, 2020, 287(12): 2577-2596.
doi: 10.1111/febs.15151 pmid: 31755197 |
| [12] | 高小晓, 孟虹, 李蓉, 等. 糖苷水解酶7家族蛋白在纤维素降解中作用的研究进展[J]. 微生物学杂志, 2020, 40(6): 113-117. |
| Gao XX, Meng H, Li R, et al. Advances in cellulose degradation by glycoside hydrolase family 7 proteins[J]. J Microbiol, 2020, 40(6): 113-117. | |
| [13] |
Taylor CB, Payne CM, Himmel ME, et al. Binding site dynamics and aromatic-carbohydrate interactions in processive and non-processive family 7 glycoside hydrolases[J]. J Phys Chem B, 2013, 117(17): 4924-4933.
doi: 10.1021/jp401410h URL |
| [14] |
Borisova AS, Eneyskaya EV, Jana S, et al. Correlation of structure, function and protein dynamics in GH7 cellobiohydrolases from Trichoderma atroviride, T. reesei and T. harzianum[J]. Biotechnol Biofuels, 2018, 11: 5.
doi: 10.1186/s13068-017-1006-7 pmid: 29344086 |
| [15] | Yang H, Shi PJ, Liu Y, et al. Loop 3 of fungal endoglucanases of glycoside hydrolase family 12 modulates catalytic efficiency[J]. Appl Environ Microbiol, 2017, 83(6): e03123-e03116. |
| [16] |
Zheng F, Tu T, Wang XY, et al. Enhancing the catalytic activity of a novel GH5 cellulase GtCel5 from Gloeophyllum trabeum CBS 900.73 by site-directed mutagenesis on loop 6[J]. Biotechnol Biofuels, 2018, 11: 76.
doi: 10.1186/s13068-018-1080-5 pmid: 29588661 |
| [17] |
Kawamoto D, Takashima T, Fukamizo T, et al. A conserved loop structure of GH19 chitinases assists the enzyme function from behind the core-functional region[J]. Glycobiology, 2022, 32(4): 356-364.
doi: 10.1093/glycob/cwab117 URL |
| [18] |
Divne C, Ståhlberg J, Teeri TT, et al. High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei[J]. J Mol Biol, 1998, 275(2): 309-325.
doi: 10.1006/jmbi.1997.1437 pmid: 9466911 |
| [19] |
von Ossowski I, Ståhlberg J, Koivula A, et al. Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A. A comparison with Phanerochaete chrysosporium Cel7D[J]. J Mol Biol, 2003, 333(4): 817-829.
doi: 10.1016/s0022-2836(03)00881-7 pmid: 14568538 |
| [20] |
Karlsson J, Momcilovic D, Wittgren B, et al. Enzymatic degradation of carboxymethyl cellulose hydrolyzed by the endoglucanases Cel5A, Cel7B, and Cel45A from Humicola insolens and Cel7B, Cel12A and Cel45Acore from Trichoderma reesei[J]. Biopolymers, 2002, 63(1): 32-40.
pmid: 11754346 |
| [21] |
Dym O, Mevarech M, Sussman JL. Structural features that stabilize halophilic malate dehydrogenase from an archaebacterium[J]. Science, 1995, 267(5202): 1344-1346.
doi: 10.1126/science.267.5202.1344 pmid: 17812611 |
| [22] |
Karan R, Mathew S, Muhammad R, et al. Understanding high-salt and cold adaptation of a polyextremophilic enzyme[J]. Microorganisms, 2020, 8(10): 1594.
doi: 10.3390/microorganisms8101594 URL |
| [23] |
Zhao B, Al Rasheed H, Ali I, et al. Efficient enzymatic saccharification of alkaline and ionic liquid-pretreated bamboo by highly active extremozymes produced by the co-culture of two halophilic fungi[J]. Bioresour Technol, 2021, 319: 124115.
doi: 10.1016/j.biortech.2020.124115 URL |
| [24] | 刘欣, 魏雪, 王凤忠, 等. 极端酶研究进展及其在食品工业中的应用现状[J]. 生物产业技术, 2017(4): 62-69. |
| Liu X, Wei X, Wang FZ, et al. Research progress of extremozymes and its application in food industry[J]. Biotechnol Bus, 2017(4): 62-69. | |
| [25] |
You S, Li J, Zhang F, et al. Loop engineering of a thermostable GH10 xylanase to improve low-temperature catalytic performance for better synergistic biomass-degrading abilities[J]. Bioresour Technol, 2021, 342: 125962.
doi: 10.1016/j.biortech.2021.125962 URL |
| [1] | YANG Jun-zhao, ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei. Structure and Function Analysis of Novel GH5 Multi-domain Cellulase [J]. Biotechnology Bulletin, 2023, 39(4): 71-80. |
| [2] | ZHANG Chen, ZHANG Tong-tong, LIU Hai-ping. Screening and Identification of Ethylene-forming Enzymes with High Activity and Thermostability [J]. Biotechnology Bulletin, 2022, 38(11): 269-276. |
| [3] | CHEN Chun, SU Ling-qia, XIA Wei, WU Jing. Improved the Thermostability of MTHase from Arthrobacter ramosus by Directed Evolution [J]. Biotechnology Bulletin, 2021, 37(3): 84-91. |
| [4] | WU Jiao, YU Gui-zhen, YUAN Hang, LIU Xian, GAO Yan-xiu, GONG Ming, ZOU Zhu-rong. Improvement on the Thermostability of Target Proteins by Fusing Rubredoxin from Hyperthermophile Pyrococcus furiosus [J]. Biotechnology Bulletin, 2021, 37(10): 110-119. |
| [5] | YUAN Lin, HUANG Zhao, ZENG Jing, GUO Jian-jun, ZHANG Ting, Lü Jun. Fusion of Phytase YiAPPA with the Raw-starch Binding Domain and Characterization of the Fusion Enzyme [J]. Biotechnology Bulletin, 2018, 34(3): 200-207. |
| [6] | ZENG Jing, GUO Jian-jun, YUAN Lin, YANG Gang, CHEN Jun. Optimization of the Thermal Activity and Stability of Hyperthermophilic α-amylase ApkA [J]. Biotechnology Bulletin, 2017, 33(8): 192-198. |
| [7] | Li Yang, Cai Haiying, Zhao Minjie, Zhang Hui, Feng Fengqin. Screening and Identification of High-yield Thermostable Lipase Producing Microorganisms [J]. Biotechnology Bulletin, 2015, 31(1): 144-150. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||