








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (3): 215-228.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0747
Previous Articles Next Articles
XIE Qian( ), JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi(
), JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi( )
)
Received:2023-08-03
Online:2024-03-26
Published:2024-04-08
Contact:
CHEN Qing-xi
E-mail:xieq0416@163.com;cqx0246@fafu.edu.cn
XIE Qian, JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi. Regulatory Genes Mining Related to Transcriptome Sequencing and Phenolic Metabolism Pathway of Canarium album Fruit with Different Fresh Food Quality[J]. Biotechnology Bulletin, 2024, 40(3): 215-228.
| 基因ID Gene ID | 引物序列Primer sequence(5'-3') | 扩增品种(系)Amplification variety(lines) |
|---|---|---|
| Cluster-0.119989-F | TGACTTCTGTGTCCTGCTCTT | DQc、TQc |
| Cluster-0.119989-R | GGTAGTGCCAACTGCTTCTTC | |
| Cluster-0.121834-F | CTTGACATGAATACAGCAGAGGAT | DQc、TQc |
| Cluster-0.121834-R | GCACCATCAATTTCAGCAACTT | |
| Cluster-0.121871-F | TCTGTGTCCTGCTCTTTGAAG | DQc、TQc |
| Cluster-0.121871-R | GCAACCTCAGAGATGGGAAG | |
| Cluster-0.121919-F | TAACCATAACGCCTCCATCTCT | DQc、TQc |
| Cluster-0.121919-R | TCATCTCACGAGACACAATAGACT | |
| Cluster-0.121949-F | TAACCATAACGCCTCCTTCTCT | DQc、TQc |
| Cluster-0.121949-R | GCTCCTACAATCACCACATCAG | |
| Cluster-0.155662-F | AGACTATGACGAGCAAGACAACT | DQc、TQc |
| Cluster-0.155662-R | GTTACCACACCACCGACTCT | |
| Cluster-0.119990-F | CTGTTGCCGTAACTTCACTGT | TQc |
| Cluster-0.119990-R | CTGAGAGATGGGAAGCGTTTC | |
| Cluster-0.120915-F | ATCATTCTCCAGGCATTTCTCAG | TQc |
| Cluster-0.120915-R | GGAACAGCCACAGCTTGTAT | |
| Cluster-0.121783-F | CAGGAAGTGTCAGCAACATCAA | TQc |
| Cluster-0.121783-R | CTGTTCTTCGCAAGGTTCATCT | |
| Cluster-0.121896-F | AGGAGTGAAGTGGTTGAAGAGTA | DQc |
| Cluster-0.121896-R | ACAATCATCCCAGGCACAATC | |
| Cluster-0.122437-F | TAACCATAACGCCTCCATCTCT | DQc |
| Cluster-0.122437-R | ATCAGTGTCCGCATGTGTAATC | |
| Cluster-0.122522-F | CCTCCATCTCTGCTTCTTCCT | DQc |
| Cluster-0.122522-R | TGTACCTGCGTGTCATCTCA | |
| 18S rRNA-F(内参基因) | CCTGAGAAACGGCTACCACA | DQc、TQc |
| 18S rRNA-R(内参基因) | CACCAGACTT GCCCTCCA |
Table 1 Primer sequences for RT-qPCR
| 基因ID Gene ID | 引物序列Primer sequence(5'-3') | 扩增品种(系)Amplification variety(lines) |
|---|---|---|
| Cluster-0.119989-F | TGACTTCTGTGTCCTGCTCTT | DQc、TQc |
| Cluster-0.119989-R | GGTAGTGCCAACTGCTTCTTC | |
| Cluster-0.121834-F | CTTGACATGAATACAGCAGAGGAT | DQc、TQc |
| Cluster-0.121834-R | GCACCATCAATTTCAGCAACTT | |
| Cluster-0.121871-F | TCTGTGTCCTGCTCTTTGAAG | DQc、TQc |
| Cluster-0.121871-R | GCAACCTCAGAGATGGGAAG | |
| Cluster-0.121919-F | TAACCATAACGCCTCCATCTCT | DQc、TQc |
| Cluster-0.121919-R | TCATCTCACGAGACACAATAGACT | |
| Cluster-0.121949-F | TAACCATAACGCCTCCTTCTCT | DQc、TQc |
| Cluster-0.121949-R | GCTCCTACAATCACCACATCAG | |
| Cluster-0.155662-F | AGACTATGACGAGCAAGACAACT | DQc、TQc |
| Cluster-0.155662-R | GTTACCACACCACCGACTCT | |
| Cluster-0.119990-F | CTGTTGCCGTAACTTCACTGT | TQc |
| Cluster-0.119990-R | CTGAGAGATGGGAAGCGTTTC | |
| Cluster-0.120915-F | ATCATTCTCCAGGCATTTCTCAG | TQc |
| Cluster-0.120915-R | GGAACAGCCACAGCTTGTAT | |
| Cluster-0.121783-F | CAGGAAGTGTCAGCAACATCAA | TQc |
| Cluster-0.121783-R | CTGTTCTTCGCAAGGTTCATCT | |
| Cluster-0.121896-F | AGGAGTGAAGTGGTTGAAGAGTA | DQc |
| Cluster-0.121896-R | ACAATCATCCCAGGCACAATC | |
| Cluster-0.122437-F | TAACCATAACGCCTCCATCTCT | DQc |
| Cluster-0.122437-R | ATCAGTGTCCGCATGTGTAATC | |
| Cluster-0.122522-F | CCTCCATCTCTGCTTCTTCCT | DQc |
| Cluster-0.122522-R | TGTACCTGCGTGTCATCTCA | |
| 18S rRNA-F(内参基因) | CCTGAGAAACGGCTACCACA | DQc、TQc |
| 18S rRNA-R(内参基因) | CACCAGACTT GCCCTCCA |
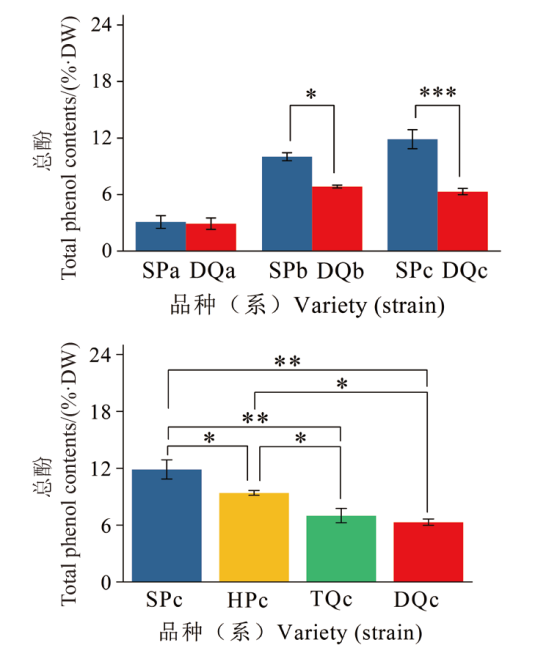
Fig. 1 Total phenol content in Chinese olive fruit ripening process and maturity period * indicates significant difference at 0.05 level, ** indicates significant difference at 0.01 level, and *** indicates significant difference at 0.001 level
| 类型Type | 序列条数Number of sequences | 序列平均长度Mean length/bp | N50/bp | N90/bp | 序列总碱基Total bases/bp |
|---|---|---|---|---|---|
| Unigene | 296 314 | 1 163 | 1 764 | 522 | 344 722 448 |
Table 2 Sequence splicing results of Chinese olive transcriptome
| 类型Type | 序列条数Number of sequences | 序列平均长度Mean length/bp | N50/bp | N90/bp | 序列总碱基Total bases/bp |
|---|---|---|---|---|---|
| Unigene | 296 314 | 1 163 | 1 764 | 522 | 344 722 448 |
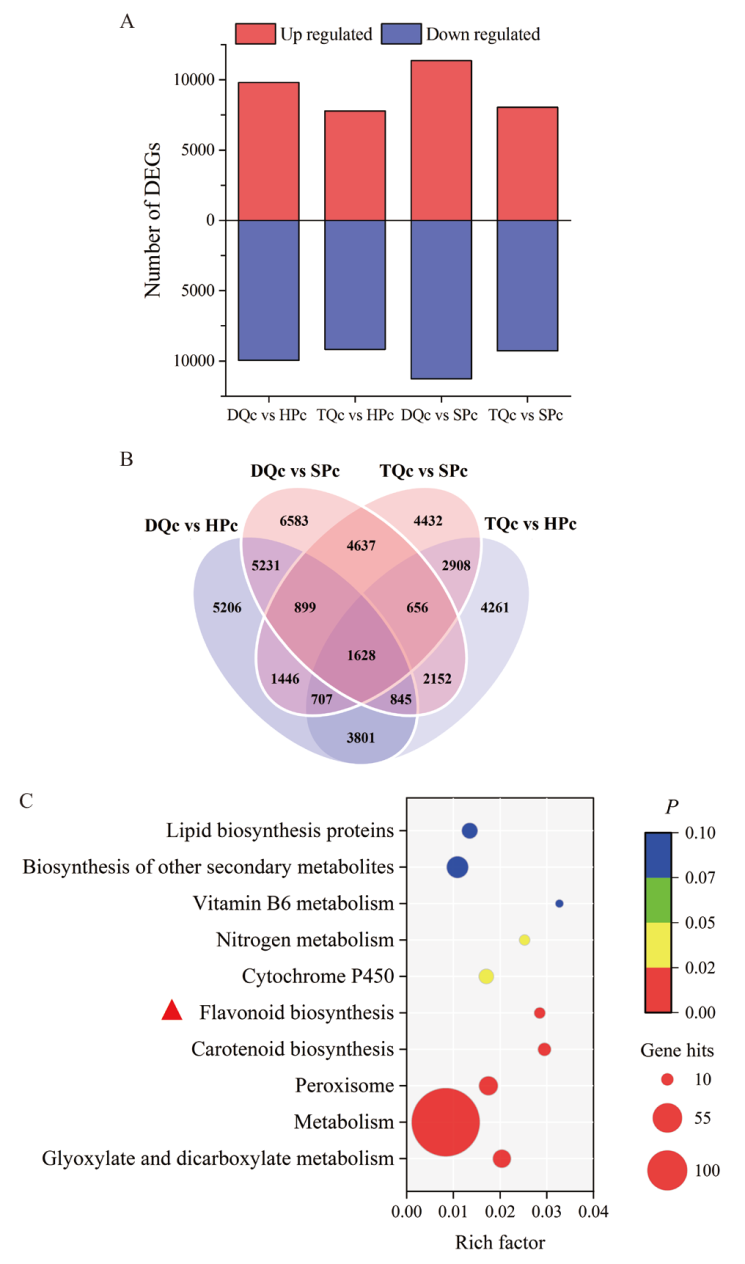
Fig. 4 Differential genes and KEGG enrichment analysis of Chinese olive in different varieties(lines) A : Up-regulated and down-regulated differential genes, gene up/down-regulated expression refers to low phenolic Chinese olive(DQc, TQc)relative to high phenolic Chinese olive(SPc, HPc); B : common or unique differential genes ;C : KEGG enrichment analysis of common differential genes

Fig. 5 Differential genes and KEGG enrichment analysis of different varieties(lines)of Chinese olive ripening process A: Differential genes in the maturation process of ‘Ziyang 1’ (SP)and ‘Dongshanchangsui’ (DQ). B : Venn of differential genes in different maturation periods. C : KEGG enrichment analysis of differential genes
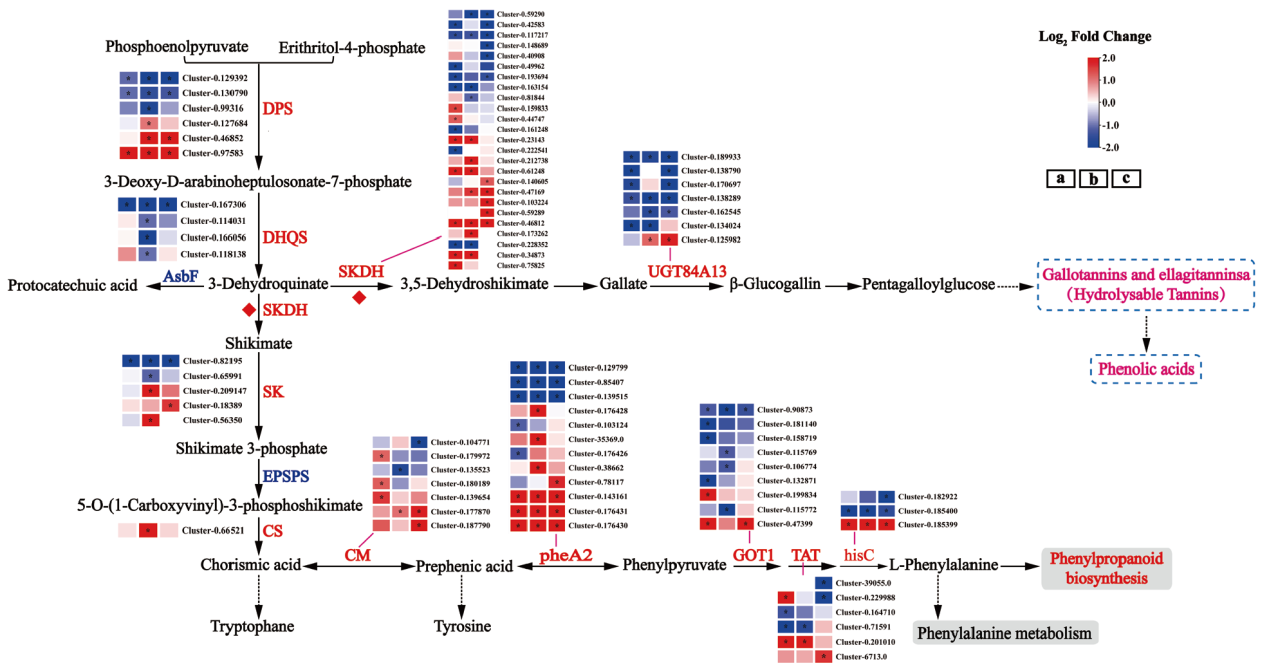
Fig. 6 Differential gene analysis of shikimic acid and hydrolyzed tannin biosynthesis pathway in different varieties(lines)of Chinese olive during maturation DPS : 3-deoxy-D-arabinoheptulose-7-phosphate synthase; DHQS : 3-dehydroquinic acid synthase; SKDH : shikimate dehydrogenase; UGTs : glycosyltransferase; SK : shikimate kinase; EPSPS : 5-enolpyruvylshikimate-3-phosphate synthase; CS : chorismate synthase; CM : chorismate mutase; pheA2 : prebenzoic acid dehydratase; GOT1 : aspartate aminotransferase; TAT : tyrosine aminotransferase; hisC : phosphohistidine aminotransferase ; Red or blue color blocks represent the value of Log2FC(DQ / SP), red and blue represent the up-regulated and down-regulated genes of low-phenol olive(DQ)compared to high-phenol olive(SP), respectively, and * indicates P.adjust < 0.05, the same below

Fig. 7 Differential gene analysis in phenylpropanes and flavonoids biosynthesis pathways in different varieties(lines)of Chinese olive during maturation PAL : Phenylalanine ammonia lyase; C4H : cinnamic acid-4-hydroxylase; 4CL : 4-coumaroyl-CoA ligase; CHS : chalcone synthase; CHI : chalcone isomerase; IFS : isoflavone synthase; FNS : flavonoid synthase; F3H : flavanone-3-hydroxylase; F3'H : flavonoid 3'-hydroxylase; F3' 5'H : flavonoid 3' 5' -hydroxylase; FLS : flavonol synthase; DFR : dihydroflavonol reductase; LAR : achromatic anthocyanin reductase; ANR : anthocyanin reductase ; aNS : anthocyanin synthase
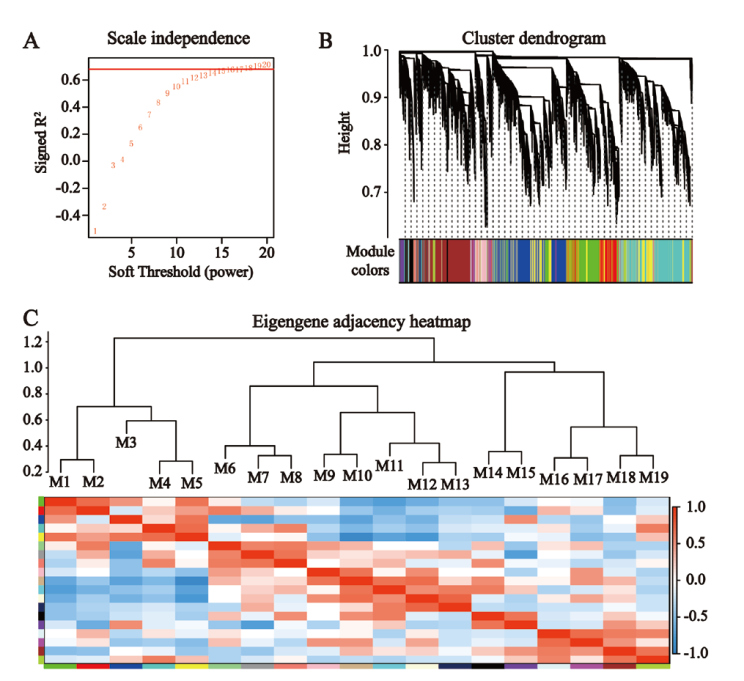
Fig. 8 Visualization of gene co-expression network A: Determination of the optimal soft threshold. B: Cluster dendrogram of co-expression modules. C: Correlation between co-expression modules

Fig. 9 Module identification of phenolic compounds and mining of their associated transcription factors A:Module and trait correlation heat map,each row corresponds to a module. The values of each lattice indicate the correlation coefficients R and P, respectively. The lattice color indicates correlation, red indicates positive correlation, and blue indicates negative correlation. B:Co-expression network analysis of phenolic synthesis genes and transcription factors. Darker colors indicate that the gene is more important in the interaction network. C:Category statistics of transcription factors related to phenol synthesis
| [1] | 陈杰忠. 果树栽培学各论: 南方本[M]. 4版. 北京: 中国农业出版社, 2011: 139-145. |
| Chen JZ. Various theories on fruit tree cultivation: southern edition[M]. 4th ed. Beijing: China Agriculture Press, 2011: 139-145. | |
| [2] | 谢倩, 李易易, 张诗艳, 等. 基于模糊数学感官评价、理化特性与电子舌的橄榄鲜食品质分析[J]. 食品科学, 2023, 44(3): 69-78. |
|
Xie Q, Li YY, Zhang SY, et al. Quality analysis of table Cauarium album L. based on fuzzy mathematics sensory evaluation, physicochemical properties and electronic tongue[J]. Food Sci, 2023, 44(3): 69-78.
doi: 10.1111/jfds.1979.44.issue-1 URL |
|
| [3] | 林玉芳, 杜正花, 陈清西. 橄榄果实品质评价因子的筛选及指标确定[J]. 热带作物学报, 2014, 35(4): 805-810. |
| Lin YF, Du ZH, Chen QX. Selection of quality evaluation indices for Chinese olive[J]. Chin J Trop Crops, 2014, 35(4): 805-810. | |
| [4] | 池毓斌, 朱丽娟, 黄敏杰, 等. 鲜食橄榄品质综合评价模型的建立与验证[J]. 果树学报, 2017, 34(8): 1051-1060. |
| Chi YB, Zhu LJ, Huang MJ, et al. Establishment and verification of a comprehensive evaluation model for quality of fresh Chinese olive[J]. J Fruit Sci, 2017, 34(8): 1051-1060. | |
| [5] | 谢倩, 张诗艳, 叶清华, 等. 鲜食橄榄发育成熟过程中多酚及相关酶活性的动态变化[J]. 果树学报, 2019, 36(6): 774-784. |
| Xie Q, Zhang SY, Ye QH, et al. Dynamic changes of polyphenols and related enzymes activity during the development and maturation of Chinese olive(Canarium album L.)[J]. J Fruit Sci, 2019, 36(6): 774-784. | |
| [6] | 池毓斌. 橄榄果实品质特性及其代谢组学的初步研究[D]. 福州: 福建农林大学, 2017. |
| Chi YB. A preliminary study on the fruit quality characteristics and metabolism of Canarium album[D]. Fuzhou: Fujian Agriculture and Forestry University, 2017. | |
| [7] | 李泽坤. 橄榄(Canarium album(lour.)raeusch)果实成熟发育蔗糖代谢变化研究[D]. 福州: 福建农林大学, 2016. |
| Li ZK. Studyies of sucrose metabolism changes in Chinese olive fruit mature development[D]. Fuzhou: Fujian Agriculture and Forestry University, 2016. | |
| [8] |
彭真汾, 叶清华, 王威, 等. 普通橄榄和清橄榄果实游离氨基酸差异成分与谷氨酰胺代谢[J]. 食品科学, 2019, 40(4): 229-236.
doi: 10.7506/spkx1002-6630-20171228-354 |
| Peng ZF, Ye QH, Wang W, et al. Differences in free amino acid composition of fruits of common olive and sweet olive and their glutamine metabolism characteristics[J]. Food Sci, 2019, 40(4): 229-236. | |
| [9] | 常强. 橄榄果实酚类物质及其抗氧化活性研究[D]. 福州: 福建农林大学, 2017. |
| Chang Q. Investigation of polyphenol compounds and their antioxidant activity in the fruits of Chinese olive[D]. Fuzhou: Fujian Agriculture and Forestry University, 2017. | |
| [10] | 谢倩. 橄榄(Canarium album(Lour.)Rauesch.)果实发育成熟过程多酚及相关酶活性研究[D]. 福州: 福建农林大学, 2014. |
| Xie Q. Polyphenol components and related enzyme activities during the Chinese olive(Canarium album(Lour.)Rauesch.)fruit development and ripening[D]. Fuzhou: Fujian Agriculture and Forestry University, 2014. | |
| [11] | 林玉芳. 福建橄榄(Canarium album(lour.)raeusch.)若干功能成分和品质相关指标的研究[D]. 福州: 福建农林大学, 2012. |
| Lin YF. Studies on some functional components and quality indexes for Chinese olive fruits in Fujian Province[D]. Fuzhou: Fujian Agriculture and Forestry University, 2012. | |
| [12] | 何志勇, 夏文水. 橄榄果实中酚类化合物的分析研究[J]. 安徽农业科学, 2008, 36(26): 11406-11407. |
| He ZY, Xia WS. Analysis of phenolic compounds in Canarium album(Lour.)raeusch fruit[J]. J Anhui Agric Sci, 2008, 36(26): 11406-11407. | |
| [13] |
蔡净蓉, 王杰, 赵俊跃, 等. 成熟期不同橄榄品种(系)果实代谢组及其差异[J]. 热带作物学报, 2022, 43(11): 2304-2315.
doi: 10.3969/j.issn.1000-2561.2022.11.015 |
| Cai JR, Wang J, Zhao JY, et al. Metabolomics and its difference of Chinese olive fruit of different varieties(lines)during the ripening period[J]. Chin J Trop Crops, 2022, 43(11): 2304-2315. | |
| [14] |
侯黔东, 沈天娇, 余欢欢, 等. 甜樱桃GH3基因家族全基因组鉴定与表达分析[J]. 园艺学报, 2021, 48(12): 2360-2374.
doi: 10.16420/j.issn.0513-353x.2020-0911 |
| Hou QD, Shen TJ, Yu HH, et al. Genome-wide identification and expression analysis of Prunus avium gretchen Hagen 3(GH3)gene family[J]. Acta Hortic Sin, 2021, 48(12): 2360-2374. | |
| [15] |
Broun P. Transcription factors as tools for metabolic engineering in plants[J]. Curr Opin Plant Biol, 2004, 7(2): 202-209.
doi: 10.1016/j.pbi.2004.01.013 pmid: 15003222 |
| [16] |
Vom Endt D, Kijne JW, Memelink J. Transcription factors controlling plant secondary metabolism: what regulates the regulators?[J]. Phytochemistry, 2002, 61(2): 107-114.
pmid: 12169302 |
| [17] |
Yang CQ, Fang X, Wu XM, et al. Transcriptional regulation of plant secondary metabolism[J]. J Integr Plant Biol, 2012, 54(10): 703-712.
doi: 10.1111/jipb.2012.54.issue-10 URL |
| [18] | 谭西北, 李鹏, 孙磊, 等. 基于WGCNA的刺葡萄抗白腐病关键基因的发掘[J]. 果树学报, 2023, 40(4): 653-668. |
| Tan XB, Li P, Sun L, et al. Discovery of key genes for White Rot resistance in Vitis davidii based on WGCNA[J]. J Fruit Sci, 2023, 40(4): 653-668. | |
| [19] |
李贵生. 猕猴桃‘金艳’和‘红阳’果实转录组的比较分析[J]. 园艺学报, 2021, 48(6): 1183-1196.
doi: 10.16420/j.issn.0513-353x.2020-0510 |
| Li GS. Comparative analysis of‘Jinyan’ and‘Hongyang’ kiwifruit transcriptomes[J]. Acta Hortic Sin, 2021, 48(6): 1183-1196. | |
| [20] |
辛建攀, 李燕, 赵楚, 等. 镉胁迫下梭鱼草叶片转录组测序及苯丙烷代谢途径相关基因挖掘[J]. 生物技术通报, 2022, 38(6): 198-210.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1151 |
| Xin JP, Li Y, Zhao C, et al. Transcriptome sequencing in the leaves of Pontederia cordata with cadmium exposure and gene mining in phenypropanoid pathways[J]. Biotechnol Bull, 2022, 38(6): 198-210. | |
| [21] | 赵权. 葡萄酚类物质及其生物合成相关结构基因表达[D]. 哈尔滨: 东北林业大学, 2010. |
| Zhao Q. Phenolic compounds and expression of related biosynthesis structural genes in grapevine[D]. Harbin: Northeast Forestry University, 2010. | |
| [22] | 林玉芳, 陈清西, 关夏玉, 等. 橄榄总多酚提取工艺优化研究[J]. 中国农学通报, 2011, 27(5): 396-400. |
| Lin YF, Chen QX, Guan XY, et al. Extraction of total polyphenol from Chinese olive(Canarium ablum L.)[J]. Chin Agric Sci Bull, 2011, 27(5): 396-400. | |
| [23] |
谢倩, 王威, 陈清西. 橄榄多酚含量测定方法的比较[J]. 食品科学, 2014, 35(8): 204-207.
doi: 10.7506/spkx1002-6630-201408040 |
| Xie Q, Wang W, Chen QX. Comparative study on three different methods for the determination of total phenolics in Chinese olive[J]. Food Sci, 2014, 35(8): 204-207. | |
| [24] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [25] |
Wang YR, Suo YJ, Han WJ, et al. Comparative transcriptomic and metabolomic analyses reveal differences in flavonoid biosynthesis between PCNA and PCA persimmon fruit[J]. Front Plant Sci, 2023, 14: 1130047.
doi: 10.3389/fpls.2023.1130047 URL |
| [26] |
Liu HX, Liu Q, Chen YL, et al. Full-length transcriptome sequencing provides insights into flavonoid biosynthesis in Camellia nitidissima Petals[J]. Gene, 2023, 850: 146924.
doi: 10.1016/j.gene.2022.146924 URL |
| [27] |
Ye QH, Zhang SY, Qiu NN, et al. Identification and characterization of glucosyltransferase that forms 1-galloyl- β-d-glucogallin in Camellia nitidissima L., a functional fruit rich in hydrolysable tannins[J]. Molecules, 2021, 26(15): 4650.
doi: 10.3390/molecules26154650 URL |
| [28] |
Liu WX, Feng Y, Yu SH, et al. The flavonoid biosynthesis network in plants[J]. Int J Mol Sci, 2021, 22(23): 12824.
doi: 10.3390/ijms222312824 URL |
| [29] | 李小溪. 葡萄果实莽草酸途径和类黄酮代谢协同调节机制的研究[D]. 北京: 中国农业大学, 2016. |
| Li XX. Collaborative expression mechanism between shikimate pathway and flavonoid metabolism[D]. Beijing: China Agricultural University, 2016. | |
| [30] |
Routaboul JM, Kerhoas L, Debeaujon I, et al. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana[J]. Planta, 2006, 224(1): 96-107.
doi: 10.1007/s00425-005-0197-5 URL |
| [31] |
Saito K, Yonekura-Sakakibara K, Nakabayashi R, et al. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity[J]. Plant Physiol Biochem, 2013, 72: 21-34.
doi: 10.1016/j.plaphy.2013.02.001 URL |
| [32] | Chen CX, Li AL. Transcriptome analysis of differentially expressed genes involved in proanthocyanidin accumulation in the rhizomes of Fagopyrum dibotrys and an irradiation-induced mutant[J]. Front Physiol, 2016, 7: 100. |
| [33] | 张小郁, 李文广, 高明堂, 等. 葡萄籽中原花青素对心肌细胞的保护作用[J]. 中药药理与临床, 2001, 17(6): 14-16. |
| Zhang XY, Li WG, Gao MT, et al. Protective effect on injured myocardial cells of proanthoc yanidins in vitro[J]. Pharmacol Clin Chin Mater Med, 2001, 17(6): 14-16. | |
| [34] |
Chen JH, Hou N, Xv X, et al. Flavonoid synthesis and metabolism during the fruit development in hickory(Carya cathayensis)[J]. Front Plant Sci, 2022, 13: 896421.
doi: 10.3389/fpls.2022.896421 URL |
| [35] |
Xia X, Gong R, Zhang CY. Integrative analysis of transcriptome and metabolome reveals flavonoid biosynthesis regulation in Rhododendron pulchrum petals[J]. BMC Plant Biol, 2022, 22(1): 401.
doi: 10.1186/s12870-022-03762-y pmid: 35974307 |
| [36] | 陈为凯. 一年两收栽培模式下葡萄果实靶向代谢组和转录组研究[D]. 北京: 中国农业大学, 2018. |
| Chen WK. Study of targeted metabolome and transcriptome in grape berries grown under double cropping viticulture system[D]. Beijing: China Agricultural University, 2018. | |
| [37] |
Jeong ST, Goto-Yamamoto N, Hashizume K, et al. Expression of the flavonoid 3'-hydroxylase and flavonoid 3', 5'-hydroxylase genes and flavonoid composition in grape(Vitis vinifera)[J]. Plant Sci, 2006, 170(1): 61-69.
doi: 10.1016/j.plantsci.2005.07.025 URL |
| [38] |
Springob K, Nakajima JI, Yamazaki M, et al. Recent advances in the biosynthesis and accumulation of anthocyanins[J]. Nat Prod Rep, 2003, 20(3): 288-303.
doi: 10.1039/b109542k pmid: 12828368 |
| [39] | 李小兰, 张明生, 吕享. 植物花青素合成酶ANS基因的研究进展[J]. 植物生理学报, 2016, 52(6): 817-827. |
| Li XL, Zhang MS, Lü X. The research progress on plant anthocyanin synthetase ANS gene[J]. Plant Physiol J, 2016, 52(6): 817-827. | |
| [40] |
Bogs J, Downey MO, Harvey JS, et al. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves[J]. Plant Physiol, 2005, 139(2): 652-663.
doi: 10.1104/pp.105.064238 pmid: 16169968 |
| [41] |
Yamasaki K, Kigawa T, Seki M, et al. DNA-binding domains of plant-specific transcription factors: structure, function, and evolution[J]. Trends Plant Sci, 2013, 18(5): 267-276.
doi: 10.1016/j.tplants.2012.09.001 pmid: 23040085 |
| [42] | 沈德绪. 果树育种学: 全国高等农业院校教材果树专业园艺专业用[M]. 第2版. 北京: 中国农业出版社, 2000. |
| Shen DX. Fruit tree breeding: textbook for fruit tree specialty and horticulture specialty in national agricultural colleges and universities[M]. 2th ed. Beijing: China Agriculture Press, 2000. | |
| [43] | 朱军. 遗传学[M]. 4版. 北京: 中国农业出版社, 2018. |
| Zhu J. Genetics[M]. 4th ed. Beijing: China Agriculture Press, 2018. | |
| [44] | 穆红梅, 杜秀菊, 张秀省, 等. 植物MYB转录因子调控苯丙烷类生物合成研究[J]. 北方园艺, 2015(24): 171-174. |
| Mu HM, Du XJ, Zhang XS, et al. Study on plants MYB transcription factors regulate biological synthesis of phenylpropanoid metabolism[J]. North Hortic, 2015(24): 171-174. | |
| [45] |
Hichri I, Barrieu F, Bogs J, et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway[J]. J Exp Bot, 2011, 62(8): 2465-2483.
doi: 10.1093/jxb/erq442 pmid: 21278228 |
| [46] |
Yoshida K, Ma DW, Constabel CP. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes[J]. Plant Physiol, 2015, 167(3): 693-710.
doi: 10.1104/pp.114.253674 pmid: 25624398 |
| [47] |
Premathilake AT, Ni JB, Bai SL, et al. R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit[J]. Planta, 2020, 252(4): 59.
doi: 10.1007/s00425-020-03473-4 |
| [48] |
Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis[J]. Trends Plant Sci, 2010, 15(10): 573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [49] |
Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J]. Plant J, 2011, 66(1): 94-116.
doi: 10.1111/tpj.2011.66.issue-1 URL |
| [50] |
孙庆国, 姜生辉, 房鸿成, 等. 苹果MdNAC9的克隆及其调控黄酮醇合成功能的鉴定[J]. 园艺学报, 2019, 46(11): 2073-2081.
doi: 10.16420/j.issn.0513-353x.2018-1070 |
| Sun QG, Jiang SH, Fang HC, et al. Cloning of MdNAC9 and functional of its regulation on flavonol synthesis[J]. Acta Hortic Sin, 2019, 46(11): 2073-2081. | |
| [51] |
Han H, Xu F, Li YT, et al. Genome-wide characterization of bZIP gene family identifies potential members involved in flavonoids biosynthesis in Ginkgo biloba L[J]. Sci Rep, 2021, 11(1): 23420.
doi: 10.1038/s41598-021-02839-2 |
| [52] |
Liu WJ, Wang YC, Yu L, et al. MdWRKY11 participates in anthocyanin accumulation in red-fleshed apples by affecting MYB transcription factors and the photoresponse factor MdHY5[J]. J Agric Food Chem, 2019, 67(32): 8783-8793.
doi: 10.1021/acs.jafc.9b02920 URL |
| [53] |
Mao ZL, Jiang HY, Wang S, et al. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple[J]. Plant Sci, 2021, 306: 110848.
doi: 10.1016/j.plantsci.2021.110848 URL |
| [54] |
An JP, Qu FJ, Yao JF, et al. Erratum: the bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple[J]. Hortic Res, 2017, 4: 17056.
doi: 10.1038/hortres.2017.56 URL |
| [55] | 罗曼. C2H2型锌指蛋白DkZF6在柿原花青素生物合成中的功能解析[D]. 武汉: 华中农业大学, 2021. |
| Luo M. Functional characterization of C2H2 type zinc finger DkZF6 in persimmon proanthocyanidins biosynthesis[D]. Wuhan: Huazhong Agricultural University, 2021. | |
| [56] |
Jamil W, Wu W, Gong H, et al. C2H2-type zinc finger proteins(DkZF1/2)synergistically control persimmon fruit deastringency[J]. Int J Mol Sci, 2019, 20(22): 5611.
doi: 10.3390/ijms20225611 URL |
| [57] | 黄萧天. 水稻Lec/B3基因调控种子休眠的作用机制的研究[D]. 自贡: 四川轻化工大学, 2021. |
| Huang XT. The mechanism of Lec/B3 genes in determination of seed dormancy in rice[D]. Zigong: Sichuan University of Science & Engineering, 2021. |
| [1] | CHEN Yan-mei. Crosstalk Between Different Post-translational Modifications and Its Regulatory Mechanisms in Plant Growth and Development [J]. Biotechnology Bulletin, 2024, 40(2): 1-8. |
| [2] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [3] | LU Yu-dan, LIU Xiao-chi, FENG Xin, CHEN Gui-xin, CHEN Yi-ting. Identification of the Kiwifruit BBX Gene Family and Analysis of Their Transcriptional Characteristics [J]. Biotechnology Bulletin, 2024, 40(2): 172-182. |
| [4] | ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses [J]. Biotechnology Bulletin, 2024, 40(2): 90-98. |
| [5] | ZHOU Hui-wen, WU Lan-hua, HAN De-peng, ZHENG Wei, YU Pao-lan, WU Yang, XIAO Xiao-jun. Genome-wide Association Study of Seed Glucosinolate Content in Brassica napus [J]. Biotechnology Bulletin, 2024, 40(1): 222-230. |
| [6] | WU Zhen, ZHANG Ming-Ying, YAN Feng, LI Yi-min, GAO Jing, YAN Yong-Gang, ZHANG Gang. Identification and Analysis of WRKY Gene Family in Rheum palmatum L. [J]. Biotechnology Bulletin, 2024, 40(1): 250-261. |
| [7] | WANG Bin, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui, HE Jin-ming. Cloning of bHLH96 Gene and Its Roles in Regulating the Biosynthesis of Peppermint Terpenes [J]. Biotechnology Bulletin, 2024, 40(1): 281-293. |
| [8] | CHEN Zhi-min, LI Cui, WEI Ji-tian, LI Xin-ran, LIU Yi, GUO Qiang. Research Progress in the Regulation of Chlorogenic Acid Biosynthesis and Its Application [J]. Biotechnology Bulletin, 2024, 40(1): 57-71. |
| [9] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [10] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [11] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [12] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [13] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [14] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [15] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||