








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 179-190.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0021
Previous Articles Next Articles
DU Bing-shuai( ), ZOU Xin-hui, WANG Zi-hao, ZHANG Xin-yuan, CAO Yi-bo, ZHANG Ling-yun(
), ZOU Xin-hui, WANG Zi-hao, ZHANG Xin-yuan, CAO Yi-bo, ZHANG Ling-yun( )
)
Received:2024-01-05
Online:2024-05-26
Published:2024-04-19
Contact:
ZHANG Ling-yun
E-mail:dubingshuai624@163.com;lyzhang@bjfu.edu.cn
DU Bing-shuai, ZOU Xin-hui, WANG Zi-hao, ZHANG Xin-yuan, CAO Yi-bo, ZHANG Ling-yun. Genome-wide Identification and Expression Analysis of the SWEET Gene Family in Camellia oleifera[J]. Biotechnology Bulletin, 2024, 40(5): 179-190.
| 基因名称Gene | 上游引物Forward primer sequence(5'-3') | 下游引物Reverse primer sequence(5'-3') |
|---|---|---|
| CoSWEET1a | GGTGAAGCCAAGAAACCTCCT | CTACTTGCTCGATCGCTTCTCT |
| CoSWEET1b | TTGGAGGAAGAGAAGCTATCTG | ATGATTTCTTGACATGCGATTGAG |
| CoSWEET2a | GCAGAGAACGCGAAAAAGGT | ATATCCAACGAAGATCTGTCGATT |
| CoSWEET2b | ATGGTTTCTTTGGCTTTGTCCC | AGATATGAGTGGCGTGCCGT |
| CoSWEET5 | CACCAGTCCCGACGTTCTTT | GGACGAAGGGAAGGCCATAG |
| CoSWEET7 | CCAAGCTCCGGTCACTCATT | ATTAGCACAGGAAGCCAGGG |
| CoSWEET9a | AGGTTCTACCAGAAGTCAAGCTG | TGGATTCGCTCGGTTTGATA |
| CoSWEET9b | AAGAGCGTGGAGTACATGCC | GCAAATCCCGAGAGACGACA |
| CoSWEET10 | TGGAGGAAGAGAAGCTATCTGAAT | TGAGGCATGACTGGAATGATTTCT |
| CoSWEET15 | TCACCACCCACTGGTGTTTAC | ACATCCAAAGCATGGCACTG |
| CoSWEET16a | TTCACCACCCACTGGTGTTT | GGCTGTGTGGTTCTTAGCCT |
| CoSWEET16b | AGACTGCAACACTAGCTGGG | TCCTCAACAGAGGACACTGC |
| CoSWEET17a | ATGTTGGGTTTCTTGGGGCA | GACAGCTAGAGGTGCTGCAT |
| CoSWEET17b | GCAAGGCCTCGATGACCAC | TTTGTCGATGCTGTATTGCCG |
| CoGAPDH | GGTGCCAAGAAGGTGGTAATA | GTTGTGCAGCTTGCATTAGAG |
| CoActin | AAACTACGGTTGCGGATAGAG | CTCCGGTGCATCCTTCATAAT |
Table 1 RT-qPCR primers used in this study
| 基因名称Gene | 上游引物Forward primer sequence(5'-3') | 下游引物Reverse primer sequence(5'-3') |
|---|---|---|
| CoSWEET1a | GGTGAAGCCAAGAAACCTCCT | CTACTTGCTCGATCGCTTCTCT |
| CoSWEET1b | TTGGAGGAAGAGAAGCTATCTG | ATGATTTCTTGACATGCGATTGAG |
| CoSWEET2a | GCAGAGAACGCGAAAAAGGT | ATATCCAACGAAGATCTGTCGATT |
| CoSWEET2b | ATGGTTTCTTTGGCTTTGTCCC | AGATATGAGTGGCGTGCCGT |
| CoSWEET5 | CACCAGTCCCGACGTTCTTT | GGACGAAGGGAAGGCCATAG |
| CoSWEET7 | CCAAGCTCCGGTCACTCATT | ATTAGCACAGGAAGCCAGGG |
| CoSWEET9a | AGGTTCTACCAGAAGTCAAGCTG | TGGATTCGCTCGGTTTGATA |
| CoSWEET9b | AAGAGCGTGGAGTACATGCC | GCAAATCCCGAGAGACGACA |
| CoSWEET10 | TGGAGGAAGAGAAGCTATCTGAAT | TGAGGCATGACTGGAATGATTTCT |
| CoSWEET15 | TCACCACCCACTGGTGTTTAC | ACATCCAAAGCATGGCACTG |
| CoSWEET16a | TTCACCACCCACTGGTGTTT | GGCTGTGTGGTTCTTAGCCT |
| CoSWEET16b | AGACTGCAACACTAGCTGGG | TCCTCAACAGAGGACACTGC |
| CoSWEET17a | ATGTTGGGTTTCTTGGGGCA | GACAGCTAGAGGTGCTGCAT |
| CoSWEET17b | GCAAGGCCTCGATGACCAC | TTTGTCGATGCTGTATTGCCG |
| CoGAPDH | GGTGCCAAGAAGGTGGTAATA | GTTGTGCAGCTTGCATTAGAG |
| CoActin | AAACTACGGTTGCGGATAGAG | CTCCGGTGCATCCTTCATAAT |
| Name | Gene ID | Chrom | CDS size | Protein length/aa | TMHs | Molecular weight(Average)/Da | pI | Instability index | GRAVY |
|---|---|---|---|---|---|---|---|---|---|
| CoSWEET1a | KAI8013233.1 | 4 | 756 | 251 | 7 | 27681.96 | 9.64 | 36.09 | 0.534 |
| CoSWEET1b | KAI8003830.1 | 9 | 726 | 241 | 7 | 26377.37 | 9.71 | 36.9 | 0.671 |
| CoSWEET2a | KAI8001534.1 | 8 | 708 | 235 | 7 | 25982.12 | 8.69 | 40.43 | 0.944 |
| CoSWEET2b | KAI8008303.1 | 7 | 708 | 235 | 7 | 25933.89 | 9.39 | 39.6 | 0.888 |
| CoSWEET5 | KAI8018837.1 | 2 | 723 | 240 | 7 | 27247.42 | 6.72 | 32.52 | 0.702 |
| CoSWEET7 | KAI7995081.1 | 12 | 771 | 256 | 7 | 27737.2 | 9.26 | 40.41 | 0.668 |
| CoSWEET9a | KAI7982657.1 | 11 | 828 | 275 | 7 | 30777.62 | 8.71 | 31.73 | 0.638 |
| CoSWEET9b | KAI7982326.1 | 11 | 831 | 276 | 6 | 31463.67 | 9.37 | 27.9 | 0.769 |
| CoSWEET10 | KAI8016974.1 | 2 | 984 | 327 | 7 | 36875.13 | 9.01 | 42.64 | 0.606 |
| CoSWEET15 | KAI8023305.1 | 6 | 894 | 297 | 7 | 33239.44 | 6.19 | 36.79 | 0.638 |
| CoSWEET16a | KAI8028642.1 | 3 | 750 | 249 | 7 | 27594.03 | 4.98 | 52.73 | 0.492 |
| CoSWEET16b | KAI7980775.1 | 1 | 789 | 262 | 3 | 28591.53 | 4.75 | 51.82 | 0.012 |
| CoSWEET17a | KAI8028641.1 | 3 | 930 | 309 | 7 | 33939.77 | 8.41 | 38.05 | 0.342 |
| CoSWEET17b | KAI7980777.1 | 1 | 909 | 302 | 7 | 33108.94 | 9.27 | 35.38 | 0.429 |
Table 2 Physical and chemical characteristics analysis of CoSWEETs family in C. oleifera
| Name | Gene ID | Chrom | CDS size | Protein length/aa | TMHs | Molecular weight(Average)/Da | pI | Instability index | GRAVY |
|---|---|---|---|---|---|---|---|---|---|
| CoSWEET1a | KAI8013233.1 | 4 | 756 | 251 | 7 | 27681.96 | 9.64 | 36.09 | 0.534 |
| CoSWEET1b | KAI8003830.1 | 9 | 726 | 241 | 7 | 26377.37 | 9.71 | 36.9 | 0.671 |
| CoSWEET2a | KAI8001534.1 | 8 | 708 | 235 | 7 | 25982.12 | 8.69 | 40.43 | 0.944 |
| CoSWEET2b | KAI8008303.1 | 7 | 708 | 235 | 7 | 25933.89 | 9.39 | 39.6 | 0.888 |
| CoSWEET5 | KAI8018837.1 | 2 | 723 | 240 | 7 | 27247.42 | 6.72 | 32.52 | 0.702 |
| CoSWEET7 | KAI7995081.1 | 12 | 771 | 256 | 7 | 27737.2 | 9.26 | 40.41 | 0.668 |
| CoSWEET9a | KAI7982657.1 | 11 | 828 | 275 | 7 | 30777.62 | 8.71 | 31.73 | 0.638 |
| CoSWEET9b | KAI7982326.1 | 11 | 831 | 276 | 6 | 31463.67 | 9.37 | 27.9 | 0.769 |
| CoSWEET10 | KAI8016974.1 | 2 | 984 | 327 | 7 | 36875.13 | 9.01 | 42.64 | 0.606 |
| CoSWEET15 | KAI8023305.1 | 6 | 894 | 297 | 7 | 33239.44 | 6.19 | 36.79 | 0.638 |
| CoSWEET16a | KAI8028642.1 | 3 | 750 | 249 | 7 | 27594.03 | 4.98 | 52.73 | 0.492 |
| CoSWEET16b | KAI7980775.1 | 1 | 789 | 262 | 3 | 28591.53 | 4.75 | 51.82 | 0.012 |
| CoSWEET17a | KAI8028641.1 | 3 | 930 | 309 | 7 | 33939.77 | 8.41 | 38.05 | 0.342 |
| CoSWEET17b | KAI7980777.1 | 1 | 909 | 302 | 7 | 33108.94 | 9.27 | 35.38 | 0.429 |
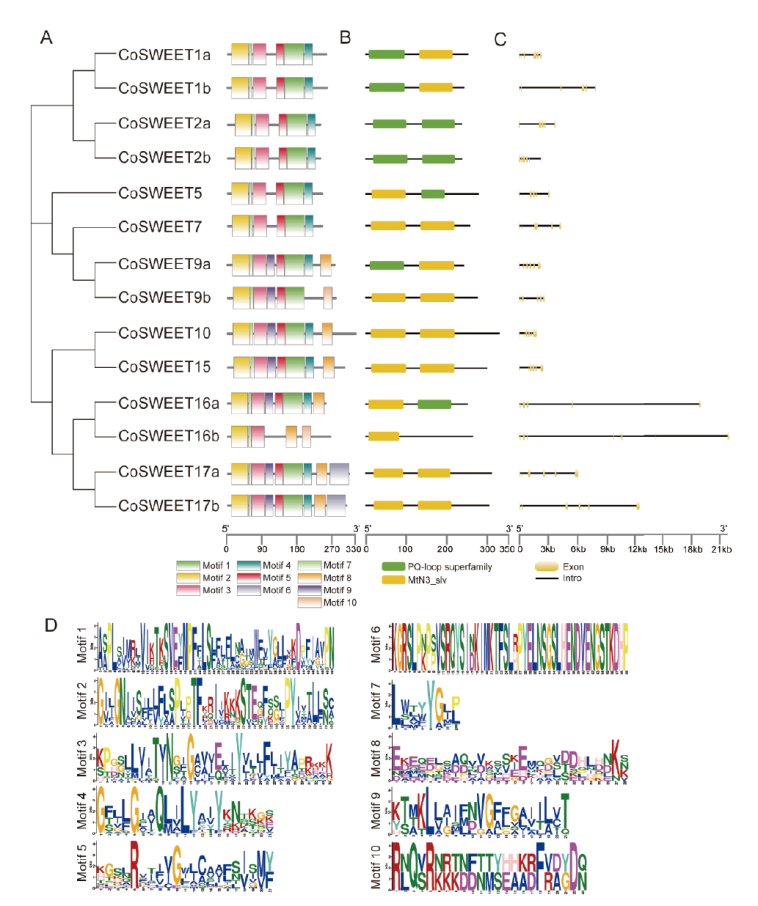
Fig. 2 Conserved motifs and exon-intron structure analysis of the CoSWEETs family in C. oleifera A: Conservative domain of CoSWEET gene family; B: conserved domain analysis of CoSWEET gene family; C: gene structure map of CoSWEET gene family; D: Logo of conserved motif of CoSWEET gene family

Fig. 4 Cis-element analysis and upstream transcription factor prediction of CoSWEET gene family in C. oleifera A: Cis-acting elements analysis in the promotors of CoSWEET gene family. B-C: Prediction about the upstream transcription factors of CoSWEET gene family
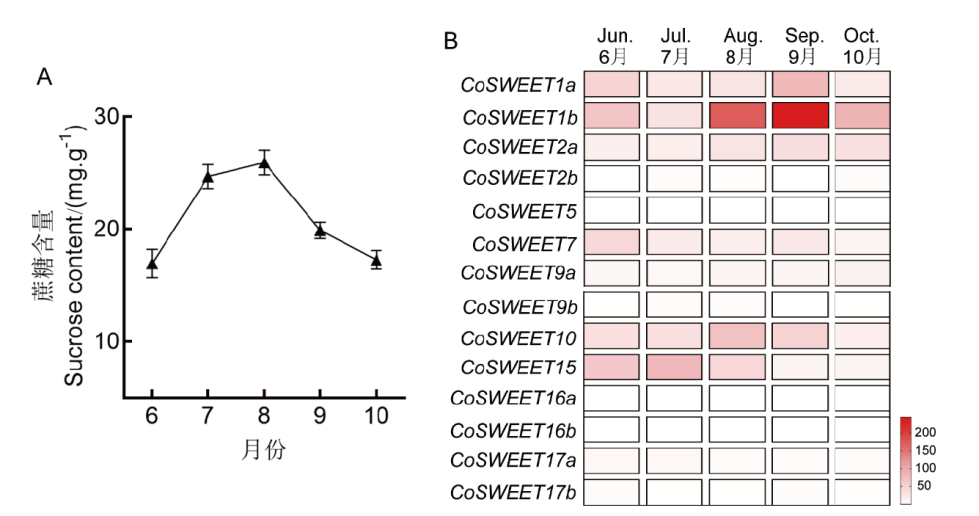
Fig. 5 Content changes of sucrose in seeds and expression analysis of CoSWEETs genes in C. oleifera A: Content changes of sucrose in seeds at different stages. B: The expression profiles of CoSWEETs in seeds at differential stages

Fig. 6 Expression analysis of CoSWEETs gene family in C. oleifera A: Expression profiles of CoSWEETs in different C. oleifera tissues. B: Expression profiles of CoSWEETs under various abiotic stress
| [1] | Ruan YL. Sucrose metabolism: gateway to diverse carbon use and sugar signaling[J]. Annu Rev Plant Biol, 2014, 65: 33-67. |
| [2] | Yoon J, Cho LH, Tun W, et al. Sucrose signaling in higher plants[J]. Plant Sci, 2021, 302: 110703. |
| [3] |
Ayre BG. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning[J]. Mol Plant, 2011, 4(3): 377-394.
doi: 10.1093/mp/ssr014 pmid: 21502663 |
| [4] |
Doidy J, Grace E, Kühn C, et al. Sugar transporters in plants and in their interactions with fungi[J]. Trends Plant Sci, 2012, 17(7): 413-422.
doi: 10.1016/j.tplants.2012.03.009 pmid: 22513109 |
| [5] | Chen LQ, Qu XQ, Hou BH, et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport[J]. Science, 2012, 335(6065): 207-211. |
| [6] | Tao YY, Cheung LS, Li S, et al. Structure of a eukaryotic SWEET transporter in a homotrimeric complex[J]. Nature, 2015, 527(7577): 259-263. |
| [7] | Xuan YH, Hu YB, Chen LQ, et al. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family[J]. Proc Natl Acad Sci USA, 2013, 110(39): E3685-E3694. |
| [8] | Chen LQ, Hou BH, Lalonde S, et al. Sugar transporters for intercellular exchange and nutrition of pathogens[J]. Nature, 2010, 468(7323): 527-532. |
| [9] |
Yuan M, Wang SP. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms[J]. Mol Plant, 2013, 6(3): 665-674.
doi: 10.1093/mp/sst035 pmid: 23430047 |
| [10] |
Gao Y, Wang ZY, Kumar V, et al. Genome-wide identification of the SWEET gene family in wheat[J]. Gene, 2018, 642: 284-292.
doi: S0378-1119(17)31009-0 pmid: 29155326 |
| [11] | Patil G, Valliyodan B, Deshmukh R, et al. Soybean(Glycine max)SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis[J]. BMC Genomics, 2015, 16(1): 520. |
| [12] | Wei XY, Liu FL, Chen C, et al. The Malus domestica sugar transporter gene family: identifications based on genome and expression profiling related to the accumulation of fruit sugars[J]. Front Plant Sci, 2014, 5: 569. |
| [13] | Jiang L, Song C, Zhu X, et al. SWEET transporters and the potential functions of these sequences in tea(Camellia sinensis)[J]. Front Genet, 2021, 12: 655843. |
| [14] | Eom JS, Chen LQ, Sosso D, et al. SWEETs, transporters for intracellular and intercellular sugar translocation[J]. Curr Opin Plant Biol, 2015, 25: 53-62. |
| [15] |
Wang SD, Yokosho K, Guo RZ, et al. The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo[J]. Plant Physiol, 2019, 180(4): 2133-2141.
doi: 10.1104/pp.19.00641 pmid: 31221732 |
| [16] | Shen S, Ma S, Chen XM, et al. A transcriptional landscape underlying sugar import for grain set in maize[J]. Plant J, 2022, 110(1): 228-242. |
| [17] | Guan YF, Huang XY, Zhu J, et al. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis[J]. Plant Physiol, 2008, 147(2): 852-863. |
| [18] | 徐磊, 王伟伟, 苏世超, 等. 小麦糖转运蛋白基因TaSWEET6的克隆与表达分析[J]. 麦类作物学报, 2016, 36(11): 1411-1418. |
| Xu L, Wang WW, Su SC, et al. Cloning and expression analysis of a sugar transporter protein gene TaSWEET6 in wheat[J]. J Triticeae Crops, 2016, 36(11): 1411-1418. | |
| [19] | 张璐, 李明, 叶广继, 等. 马铃薯糖转运蛋白基因的克隆及表达分析[J]. 西北植物学报, 2019, 39(9): 1528-1533. |
| Zhang L, Li M, Ye GJ, et al. Cloning and expression analysis of a sugar transporter protein gene in potato[J]. Acta Bot Boreali Occidentalia Sin, 2019, 39(9): 1528-1533. | |
| [20] | 马来. 水稻蔗糖转运蛋白OsSWEET11和OsSWEET14功能的研究[D]. 南京: 南京农业大学, 2018. |
| Ma L. Functional analysis of sucrose transporter genes OsSWEET11 and OsSWEET14 in rice[D]. Nanjing: Nanjing Agricultural University, 2018. | |
| [21] | Durand M, Porcheron B, Hennion N, et al. Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots[J]. Plant Physiol, 2016, 170(3): 1460-1479. |
| [22] |
Valifard M, Le Hir R, Müller J, et al. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance[J]. Plant Physiol, 2021, 187(4): 2716-2730.
doi: 10.1093/plphys/kiab436 pmid: 34597404 |
| [23] |
叶洲辰, 吴友根, 于靖, 等. 不同产地油茶籽油提取物的抗氧化活性比较分析及其营养评价[J]. 生物技术通报, 2019, 35(10): 80-88.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0433 |
| Ye ZC, Wu YG, Yu J, et al. Comparative analysis of antioxidant activities of Camellia oleifera oil extracts from different areas and their nutritional assessments[J]. Biotechnol Bull, 2019, 35(10): 80-88. | |
| [24] | Zhang FH, Li Z, Zhou JQ, et al. Comparative study on fruit development and oil synthesis in two cultivars of Camellia oleifera[J]. BMC Plant Biol, 2021, 21(1): 348. |
| [25] | Song QL, Gong WF, Yu XR, et al. Transcriptome and anatomical comparisons reveal the effects of methyl jasmonate on the seed development of Camellia oleifera[J]. J Agric Food Chem, 2023, 71(17): 6747-6762. |
| [26] | He ZL, Liu CX, Zhang Z, et al. Integration of mRNA and miRNA analysis reveals the differentially regulatory network in two different Camellia oleifera cultivars under drought stress[J]. Front Plant Sci, 2022, 13: 1001357. |
| [27] | Wu YT, Wu PZ, Xu SM, et al. Genome-wide identification, expression patterns and sugar transport of the physic nut SWEET gene family and a functional analysis of JcSWEET16 in Arabidopsis[J]. Int J Mol Sci, 2022, 23(10): 5391. |
| [28] | Hu Z, Tang ZJ, Zhang YM, et al. Rice SUT and SWEET transporters[J]. Int J Mol Sci, 2021, 22(20): 11198. |
| [29] | Zeng Z, Lyu T, Lyu YM. LoSWEET14, a sugar transporter in lily, is regulated by transcription factor LoABF2 to participate in the ABA signaling pathway and enhance tolerance to multiple abiotic stresses in tobacco[J]. Int J Mol Sci, 2022, 23(23): 15093. |
| [30] | Yu MX, Liu JL, Du BS, et al. NAC transcription factor PwNAC11 activates ERD1 by interaction with ABF3 and DREB2A to enhance drought tolerance in transgenic Arabidopsis[J]. Int J Mol Sci, 2021, 22(13): 6952. |
| [31] | Dai ZR, Yan PY, He SZ, et al. Genome-wide identification and expression analysis of SWEET family genes in sweet potato and its two diploid relatives[J]. Int J Mol Sci, 2022, 23(24): 15848. |
| [32] |
孙全喜, 苑翠玲, 牟艺菲, 等. 花生SWEET基因全基因组鉴定及表达分析[J]. 作物学报, 2023, 49(4): 938-954.
doi: 10.3724/SP.J.1006.2023.24066 |
| Sun QX, Yuan CL, Mou YF, et al. Genome-wide identification and expression analysis of SWEET genes from peanut genomes[J]. Acta Agron Sin, 2023, 49(4): 938-954. | |
| [33] |
Yang J, Luo DP, Yang B, et al. SWEET11 and 15 as key players in seed filling in rice[J]. New Phytol, 2018, 218(2): 604-615.
doi: 10.1111/nph.15004 pmid: 29393510 |
| [34] |
Saddhe AA, Manuka R, Penna S. Plant sugars: Homeostasis and transport under abiotic stress in plants[J]. Physiol Plant, 2021, 171(4): 739-755.
doi: 10.1111/ppl.13283 pmid: 33215734 |
| [35] | Wu YT, Wang SS, Du WH, et al. Sugar transporter ZmSWEET1b is responsible for assimilate allocation and salt stress response in maize[J]. Funct Integr Genomics, 2023, 23(2): 137. |
| [36] |
路静, 马齐军, 康慧, 等. 苹果糖转运蛋白基因MdSWEET1在番茄中异源表达提高其耐盐性[J]. 园艺学报, 2019, 46(3): 433-443.
doi: 10.16420/j.issn.0513-353x.2018-0744 |
| Lu J, Ma QJ, Kang H, et al. Ectopic expressing MdSWEET1 in tomato enhanced salt tolerance[J]. Acta Hortic Sin, 2019, 46(3): 433-443. | |
| [37] | Ye ZH, Du BS, Zhou J, et al. Camellia oleifera CoSWEET10 is crucial for seed development and drought resistance by mediating su-gar transport in transgenic Arabidopsis[J]. Plants, 2023, 12(15): 2818. |
| [38] | 胡晓波. 柑橘SWEET11d对果实蔗糖积累的转录调控机制研究[D]. 杭州: 浙江大学, 2022. |
| Hu XB. Transcriptional regulation mechanism of citrus SWEET11d on fruit sucrose accumulation[D]. Hangzhou: Zhejiang University, 2022. | |
| [39] | Wu YF, Lee SK, Yoo Y, et al. Rice transcription factor OsDOF11 modulates sugar transport by promoting expression of sucrose transporter and SWEET genes[J]. Mol Plant, 2018, 11(6): 833-845. |
| [40] | Sun WJ, Gao ZY, Wang J, et al. Cotton fiber elongation requires the transcription factor GhMYB212 to regulate transportation into expanding fibers[J]. New Phytol, 2019, 222(2): 864-881. |
| [1] | GUO Hui-yan, DONG Xue, AN Meng-nan, XIA Zi-hao, WU Yuan-hua. Research Progress in the Functions of Key Enzymes of Ubiquitination Modification in Plant Stress Responses [J]. Biotechnology Bulletin, 2024, 40(4): 1-11. |
| [2] | PENG Feng, YU Hai-xia, ZHANG Kun, LIU Ying-ying, TAN Gui-yu. Review on the Regulation of Caleosin on Plant Lipid Droplet [J]. Biotechnology Bulletin, 2024, 40(4): 33-39. |
| [3] | JIANG Lin-qi, ZHAO Jia-ying, ZHENG Fei-xiong, YAO Xin-yi, LI Xiao-xian, YU Zhen-ming. Identification and Expression Analysis of 14-3-3 Gene Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2024, 40(3): 229-241. |
| [4] | ZHOU Hong-dan, LUO Xiao-ping, TU Mi-xue, LI Zhong-guang. Phytomelatonin: An Emerging Signal Molecule Responding to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 41-51. |
| [5] | WU Cui-cui, XIAO Shui-ping. Genome-wide Identification of HD-Zip Gene Family in Gossypium hirsutum L. and Expression Analysis in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(2): 130-145. |
| [6] | XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane [J]. Biotechnology Bulletin, 2024, 40(2): 197-211. |
| [7] | LI Ya-nan, ZHANG Hao-jie, LIANG Meng-jing, LUO Tao, LI Wang-ning, ZHANG Chun-hui, JI Chun-li, LI Run-zhi, XUE Jin-ai, CUI Hong-li. Identification and Expression Analysis of Calcium-dependent Protein Kinase(CDPK)Family in Haematococcus pluvialis [J]. Biotechnology Bulletin, 2024, 40(2): 300-312. |
| [8] | ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses [J]. Biotechnology Bulletin, 2024, 40(2): 90-98. |
| [9] | ZHANG Yi, ZHANG Xin-ru, ZHANG Jin-ke, HU Li-zong, SHANGGUAN Xin-xin, ZHENG Xiao-hong, HU Juan-juan, ZHANG Cong-cong, MU Gui-qing, LI Cheng-wei. Functional Analysis of TaMYB1 Gene in Wheat Under Cadmium Stress [J]. Biotechnology Bulletin, 2024, 40(1): 194-206. |
| [10] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [11] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [12] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [13] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [14] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [15] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||