








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 134-142.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0066
Previous Articles Next Articles
BAI Zhi-yuan1( ), XU Fei1,2, YANG Wu1,2, WANG Ming-gui1,2, YANG Yu-hua1, ZHANG Hai-ping1,2(
), XU Fei1,2, YANG Wu1,2, WANG Ming-gui1,2, YANG Yu-hua1, ZHANG Hai-ping1,2( ), ZHANG Rui-jun1,2(
), ZHANG Rui-jun1,2( )
)
Received:2024-01-16
Online:2024-06-26
Published:2024-05-14
Contact:
ZHANG Hai-ping, ZHANG Rui-jun
E-mail:bzy923@163.com;nkyzhp@126.com;zrj013835@163.com
BAI Zhi-yuan, XU Fei, YANG Wu, WANG Ming-gui, YANG Yu-hua, ZHANG Hai-ping, ZHANG Rui-jun. Transcriptome Analysis of Fertility Transformation in Weakly Restoring Hybrid F1 of Soybean Cytoplasmic Male Sterility[J]. Biotechnology Bulletin, 2024, 40(6): 134-142.
| 基因ID Gene_id | 基因注释 Gene annotation | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|---|
| AFR34332.1 | COX2 | TGGCTATTCCTCACGATTGCT | GCATCATAGGTGTTGCTGCG |
| AFR34381.1 | ORF103c | AAACTCGAAAGGGTGGAAGTCT | TCCCCTGCATAGGGAATACCA |
| AFR34382.1 | ORF178 | GGCATCGGTCCGTAGAACTT | TCCATAACATAACGGGGCGG |
| Glyma.02G008300 | 果胶甲基酯酶Pectin methylesterase | CTCATTAGCAAGCAGGAGGCT | AGCATCGAAAGGAACGCCAG |
| Glyma.13G064700 | 果胶裂解酶Pectin lyase | CGACAACGCCAAGGAGATCA | ATGGGTGAGCCTGAAGTGAC |
| Glyma.07G246600 | UDP葡萄糖醛酸脱羧酶 UDP glucuronic acid decarboxylase | AACCGCACCTCAGATTCACC | TGAGGAGCCGACACTATGGA |
| Glyma.20G103900 | 糖转运蛋白11 Sugar transporter protein 11 | AGCTTGTTCTTGCAAGTGAGTC | CTGAGGCCTGTATTTTGGCG |
| Glyma.02G154400 | 谷胱甘肽S转移酶Glutathione S-transferase | CTGCATCACCTCCCAACGAT | ATATCAGCCACCCATGCACC |
| Glyma.07G225400 | L抗坏血酸氧化酶L-ascorbic acid oxidase | GTGCCCCAGCAGGTTATTCT | CCCAATGGAAAAGCAAGGGC |
| Glyma.01G011300 | 含五肽重复序列的蛋白质At1g31920 PPR At1g31920 | GACCGTAATCGCTTCCACCA | AAGATGCTCAACATACCACAATG |
| Glyma.08G038200 | 含五肽重复序列的蛋白质At5g21222异构体X1 PPR At5g21222 isoform X1 | TGGGGATCACTTCTTGGTGC | TCAGTTTCCTGACCTGACGC |
Table 1 11 related genes and their primer sequences
| 基因ID Gene_id | 基因注释 Gene annotation | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|---|
| AFR34332.1 | COX2 | TGGCTATTCCTCACGATTGCT | GCATCATAGGTGTTGCTGCG |
| AFR34381.1 | ORF103c | AAACTCGAAAGGGTGGAAGTCT | TCCCCTGCATAGGGAATACCA |
| AFR34382.1 | ORF178 | GGCATCGGTCCGTAGAACTT | TCCATAACATAACGGGGCGG |
| Glyma.02G008300 | 果胶甲基酯酶Pectin methylesterase | CTCATTAGCAAGCAGGAGGCT | AGCATCGAAAGGAACGCCAG |
| Glyma.13G064700 | 果胶裂解酶Pectin lyase | CGACAACGCCAAGGAGATCA | ATGGGTGAGCCTGAAGTGAC |
| Glyma.07G246600 | UDP葡萄糖醛酸脱羧酶 UDP glucuronic acid decarboxylase | AACCGCACCTCAGATTCACC | TGAGGAGCCGACACTATGGA |
| Glyma.20G103900 | 糖转运蛋白11 Sugar transporter protein 11 | AGCTTGTTCTTGCAAGTGAGTC | CTGAGGCCTGTATTTTGGCG |
| Glyma.02G154400 | 谷胱甘肽S转移酶Glutathione S-transferase | CTGCATCACCTCCCAACGAT | ATATCAGCCACCCATGCACC |
| Glyma.07G225400 | L抗坏血酸氧化酶L-ascorbic acid oxidase | GTGCCCCAGCAGGTTATTCT | CCCAATGGAAAAGCAAGGGC |
| Glyma.01G011300 | 含五肽重复序列的蛋白质At1g31920 PPR At1g31920 | GACCGTAATCGCTTCCACCA | AAGATGCTCAACATACCACAATG |
| Glyma.08G038200 | 含五肽重复序列的蛋白质At5g21222异构体X1 PPR At5g21222 isoform X1 | TGGGGATCACTTCTTGGTGC | TCAGTTTCCTGACCTGACGC |
| 样品 Sample | Reads总数 Total reads | 碱基总数 Total bases | Q20/% | Q30/% | 过滤读数 Clean reads | 过滤碱基数Clean bases | 过滤数百分比 Clean percentage/% | 比对总数 Total_ mapped | 比对总数百分比Total_ mapped percentage/% | 单个比对数 Uniquely_mapped | 单个比对数百分比Uniquely_mapped percentage/% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A_T_1 | 51 129 282 | 7 669 392 300 | 98.01 | 94.68 | 47 150 672 | 7 072 600 800 | 92.21 | 45 134 613 | 95.72 | 44 178 583 | 97.88 |
| A_T_2 | 51 432 310 | 7 714 846 500 | 98.03 | 94.73 | 47 431 430 | 7 114 714 500 | 92.22 | 45 474 740 | 95.87 | 44 424 415 | 97.69 |
| A_T_3 | 43 683 610 | 6 552 541 500 | 97.93 | 94.56 | 40 201 490 | 6 030 223 500 | 92.02 | 38 478 493 | 95.71 | 37 620 308 | 97.77 |
| A_CK_1 | 51 196 992 | 7 679 548 800 | 97.98 | 94.65 | 47 273 686 | 7 091 052 900 | 92.33 | 45 349 845 | 95.93 | 44 350 422 | 97.80 |
| A_CK_2 | 46 737 644 | 7 010 646 600 | 98.18 | 94.95 | 43 318 284 | 6 497 742 600 | 92.68 | 41 632 540 | 96.11 | 40 757 276 | 97.90 |
| A_CK_3 | 49 420 632 | 7 413 094 800 | 98.05 | 94.75 | 45 611 544 | 6 841 731 600 | 92.29 | 43 802 214 | 96.03 | 42 854 974 | 97.84 |
| 平均 Average | 48 933 412 | 7 340 011 750 | 98.03 | 94.72 | 45 164 518 | 6 774 677 650 | 92.29 | 43 312 074 | 95.90 | 42 364 330 | 97.81 |
Table 2 Sorting, filtering and comparison statistics of transcriptome sequencing data
| 样品 Sample | Reads总数 Total reads | 碱基总数 Total bases | Q20/% | Q30/% | 过滤读数 Clean reads | 过滤碱基数Clean bases | 过滤数百分比 Clean percentage/% | 比对总数 Total_ mapped | 比对总数百分比Total_ mapped percentage/% | 单个比对数 Uniquely_mapped | 单个比对数百分比Uniquely_mapped percentage/% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A_T_1 | 51 129 282 | 7 669 392 300 | 98.01 | 94.68 | 47 150 672 | 7 072 600 800 | 92.21 | 45 134 613 | 95.72 | 44 178 583 | 97.88 |
| A_T_2 | 51 432 310 | 7 714 846 500 | 98.03 | 94.73 | 47 431 430 | 7 114 714 500 | 92.22 | 45 474 740 | 95.87 | 44 424 415 | 97.69 |
| A_T_3 | 43 683 610 | 6 552 541 500 | 97.93 | 94.56 | 40 201 490 | 6 030 223 500 | 92.02 | 38 478 493 | 95.71 | 37 620 308 | 97.77 |
| A_CK_1 | 51 196 992 | 7 679 548 800 | 97.98 | 94.65 | 47 273 686 | 7 091 052 900 | 92.33 | 45 349 845 | 95.93 | 44 350 422 | 97.80 |
| A_CK_2 | 46 737 644 | 7 010 646 600 | 98.18 | 94.95 | 43 318 284 | 6 497 742 600 | 92.68 | 41 632 540 | 96.11 | 40 757 276 | 97.90 |
| A_CK_3 | 49 420 632 | 7 413 094 800 | 98.05 | 94.75 | 45 611 544 | 6 841 731 600 | 92.29 | 43 802 214 | 96.03 | 42 854 974 | 97.84 |
| 平均 Average | 48 933 412 | 7 340 011 750 | 98.03 | 94.72 | 45 164 518 | 6 774 677 650 | 92.29 | 43 312 074 | 95.90 | 42 364 330 | 97.81 |
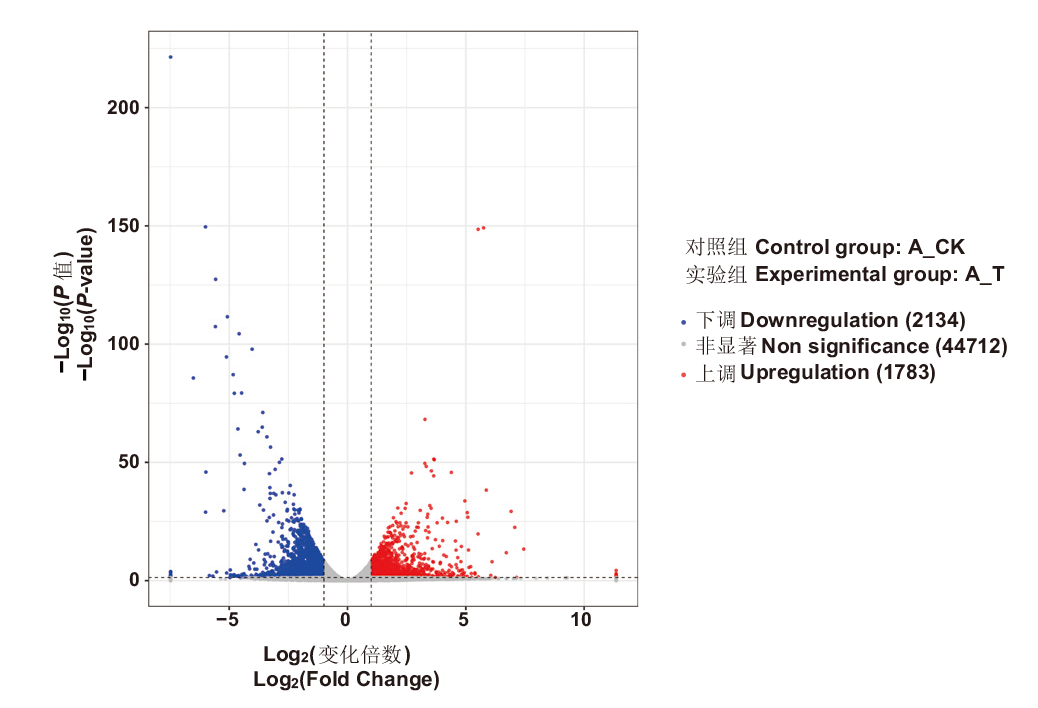
Fig. 1 Volcano map of differentially expressed genes during the fertility transformation of weakly restoring cytoplasmic male sterile hybrid F1(H3A×SXTH3)in soybean Blue dots indicate downregulation of differentially expressed genes, red dots indicate upregulation of differentially expressed genes, and gray dots indicated non-significance of differentially expressed genes
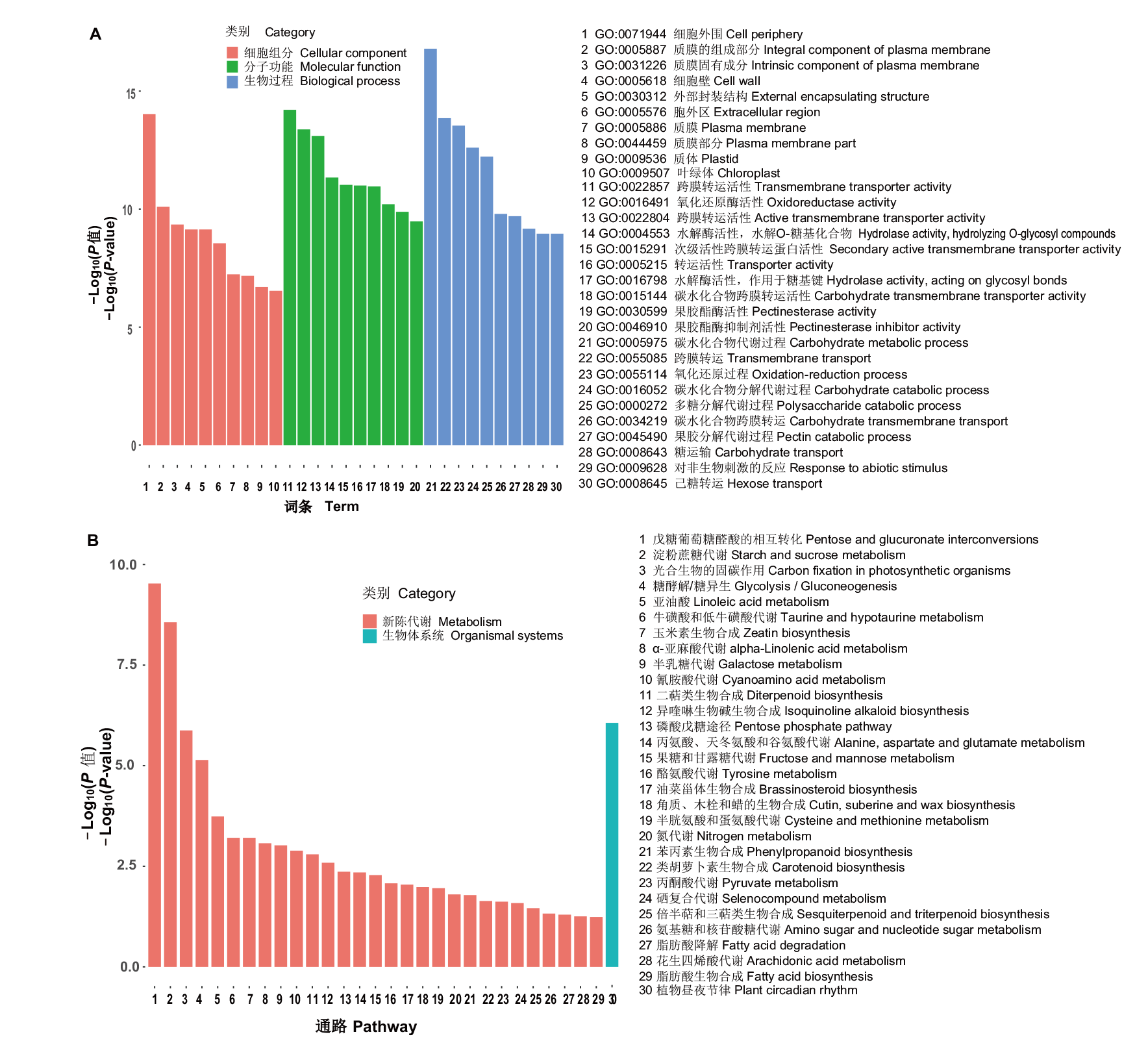
Fig. 2 Functional enrichment analysis of differentially expressed genes during the fertility transformation of weakly restoring cytoplasmic male sterile hybrid F1(H3A×SXTH3)in soybean A: GO enrichment analysis of differentially expressed genes. B: KEGG enrichment analysis of differentially expressed genes
| 基因ID Gene ID | A_CK平均值 baseMean_A_CK | A_T平均值 baseMean_A_T | 基因注释 Gene annotation | Log2(变化倍数) log2(FC) | P值 P value |
|---|---|---|---|---|---|
| Glyma.03G261800 | 9 951.556 4 | 55.757 5 | MYB转录因子MYB114 MYB transcription factor MYB114 | -7.479 6 | 4.09E-222 |
| Glyma.19G260900 | 3 490.330 4 | 54.321 4 | LHY蛋白 LHY protein | -6.005 7 | 2.61E-150 |
| Glyma.16G017400 | 14 705.494 2 | 897.924 7 | 晚期伸长下胚轴/昼夜节律时钟相关蛋白 Late elongation hypocotyl/ circadian clock related proteins | -4.033 6 | 1.45E-98 |
| Glyma.02G267800 | 662.352 9 | 165.158 2 | E3泛素蛋白连接酶COP1 E3 ubiquitin-protein ligase COP1 | -2.003 8 | 5.92E-20 |
| Glyma.14G049700 | 1 034.788 7 | 263.650 1 | E3泛素蛋白连接酶COP1 E3 ubiquitin-protein ligase COP1 | -1.972 6 | 2.56E-22 |
| Glyma.07G048500 | 1 425.609 8 | 402.471 1 | LHY1蛋白 LHY1 protein | -1.824 6 | 5.89E-21 |
| Glyma.07G013500 | 264.329 0 | 87.150 1 | ELF3蛋白2亚型X1 ELF3 protein 2 isoform X1 | -1.600 8 | 1.67E-09 |
| Glyma.03G225000 | 801.462 1 | 309.803 6 | 转录因子PIF3 Transcription factor PIF3 | -1.371 3 | 9.81E-12 |
| Glyma.14G174200 | 959.094 4 | 386.228 3 | 隐色素-1 Cryptochrome-1 | -1.312 2 | 2.63E-11 |
| Glyma.16G092700 | 205.123 5 | 102.488 5 | bZIP转录因子bZIP69 bZIP transcription factor bZIP69 | -1.001 0 | 0.000 170 8 |
| Glyma.04G228300 | 520.821 0 | 5 515.352 4 | 双组分响应调节器样APRR9亚型X2 Two component response regulator like APRR9 subtype X2 | 3.404 6 | 1.95E-23 |
| Glyma.16G150700 | 1.939 3 | 16.919 3 | 磷脂酰乙醇胺结合蛋白FT2a Phosphatidyl ethanolamine binding protein FT2a | 3.125 1 | 0.043 659 5 |
| Glyma.06G136600 | 232.560 3 | 1 063.196 7 | 双组分响应调节器,如PRR95 Two component response regulator-like PRR95 | 2.192 7 | 3.16E-05 |
| Glyma.07G058200 | 340.731 0 | 1 475.765 4 | PHYA-105 1亚型X2的蛋白抑制剂 Protein inhibitor of PHYA-105 1 isoform X2 | 2.114 8 | 4.15E-05 |
| Glyma.10G221500 | 2 158.991 6 | 6 635.026 4 | GIGANTEA核蛋白 GIGANTEA nuclear protein | 1.619 7 | 4.07E-10 |
| Glyma.16G027200 | 342.509 0 | 880.504 9 | PHYA-105蛋白抑制剂 Protein suppressor of PHYA-105 | 1.362 2 | 7.15E-12 |
| Glyma.01G192100 | 426.999 5 | 1 089.072 6 | 酪蛋白激酶II亚单位α-2 Casein kinase II subunit α-2 | 1.350 8 | 2.99E-12 |
| Glyma.04G050200 | 542.052 4 | 1 120.864 8 | 早花相关蛋白 Ealrly flower related proteins | 1.048 1 | 0.002 186 9 |
| Glyma.19G260400 | 586.539 7 | 1 207.006 2 | 双组分响应调节器样PRR95亚型X1 Two component response regulator-like PRR95 isoform X1 | 1.041 1 | 0.018 590 2 |
| Glyma.13G135900 | 1 456.119 6 | 2 977.895 5 | 双组分响应调节器APRR7亚型X1 Two component response regulator-like APRR7 isoform X1 | 1.032 2 | 4.15E-09 |
Table 3 Differentially expressed genes analysis in plant circadian rhythm of KEGG pathway during the fertility transformation of weakly restoring cytoplasmic male sterile hybrid F1(H3A×SXTH3)in soybean
| 基因ID Gene ID | A_CK平均值 baseMean_A_CK | A_T平均值 baseMean_A_T | 基因注释 Gene annotation | Log2(变化倍数) log2(FC) | P值 P value |
|---|---|---|---|---|---|
| Glyma.03G261800 | 9 951.556 4 | 55.757 5 | MYB转录因子MYB114 MYB transcription factor MYB114 | -7.479 6 | 4.09E-222 |
| Glyma.19G260900 | 3 490.330 4 | 54.321 4 | LHY蛋白 LHY protein | -6.005 7 | 2.61E-150 |
| Glyma.16G017400 | 14 705.494 2 | 897.924 7 | 晚期伸长下胚轴/昼夜节律时钟相关蛋白 Late elongation hypocotyl/ circadian clock related proteins | -4.033 6 | 1.45E-98 |
| Glyma.02G267800 | 662.352 9 | 165.158 2 | E3泛素蛋白连接酶COP1 E3 ubiquitin-protein ligase COP1 | -2.003 8 | 5.92E-20 |
| Glyma.14G049700 | 1 034.788 7 | 263.650 1 | E3泛素蛋白连接酶COP1 E3 ubiquitin-protein ligase COP1 | -1.972 6 | 2.56E-22 |
| Glyma.07G048500 | 1 425.609 8 | 402.471 1 | LHY1蛋白 LHY1 protein | -1.824 6 | 5.89E-21 |
| Glyma.07G013500 | 264.329 0 | 87.150 1 | ELF3蛋白2亚型X1 ELF3 protein 2 isoform X1 | -1.600 8 | 1.67E-09 |
| Glyma.03G225000 | 801.462 1 | 309.803 6 | 转录因子PIF3 Transcription factor PIF3 | -1.371 3 | 9.81E-12 |
| Glyma.14G174200 | 959.094 4 | 386.228 3 | 隐色素-1 Cryptochrome-1 | -1.312 2 | 2.63E-11 |
| Glyma.16G092700 | 205.123 5 | 102.488 5 | bZIP转录因子bZIP69 bZIP transcription factor bZIP69 | -1.001 0 | 0.000 170 8 |
| Glyma.04G228300 | 520.821 0 | 5 515.352 4 | 双组分响应调节器样APRR9亚型X2 Two component response regulator like APRR9 subtype X2 | 3.404 6 | 1.95E-23 |
| Glyma.16G150700 | 1.939 3 | 16.919 3 | 磷脂酰乙醇胺结合蛋白FT2a Phosphatidyl ethanolamine binding protein FT2a | 3.125 1 | 0.043 659 5 |
| Glyma.06G136600 | 232.560 3 | 1 063.196 7 | 双组分响应调节器,如PRR95 Two component response regulator-like PRR95 | 2.192 7 | 3.16E-05 |
| Glyma.07G058200 | 340.731 0 | 1 475.765 4 | PHYA-105 1亚型X2的蛋白抑制剂 Protein inhibitor of PHYA-105 1 isoform X2 | 2.114 8 | 4.15E-05 |
| Glyma.10G221500 | 2 158.991 6 | 6 635.026 4 | GIGANTEA核蛋白 GIGANTEA nuclear protein | 1.619 7 | 4.07E-10 |
| Glyma.16G027200 | 342.509 0 | 880.504 9 | PHYA-105蛋白抑制剂 Protein suppressor of PHYA-105 | 1.362 2 | 7.15E-12 |
| Glyma.01G192100 | 426.999 5 | 1 089.072 6 | 酪蛋白激酶II亚单位α-2 Casein kinase II subunit α-2 | 1.350 8 | 2.99E-12 |
| Glyma.04G050200 | 542.052 4 | 1 120.864 8 | 早花相关蛋白 Ealrly flower related proteins | 1.048 1 | 0.002 186 9 |
| Glyma.19G260400 | 586.539 7 | 1 207.006 2 | 双组分响应调节器样PRR95亚型X1 Two component response regulator-like PRR95 isoform X1 | 1.041 1 | 0.018 590 2 |
| Glyma.13G135900 | 1 456.119 6 | 2 977.895 5 | 双组分响应调节器APRR7亚型X1 Two component response regulator-like APRR7 isoform X1 | 1.032 2 | 4.15E-09 |
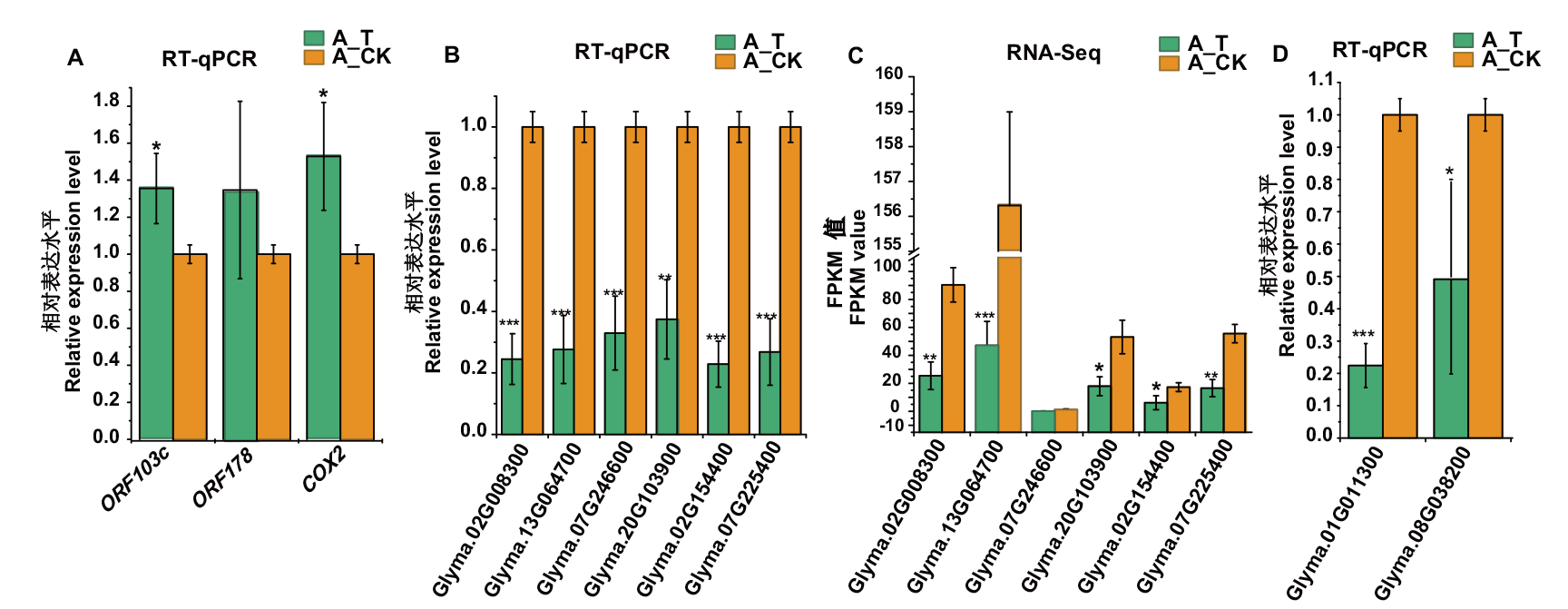
Fig. 3 RT-qPCR and RNA-seq analysis of selected differentially expressed genes during the fertility transformation of weakly restoring cytoplasmic male sterile hybrid F1(H3A×SXTH3)in soybean A: RT-qPCR analysis of mitochondrial genes related to cytoplasmic male sterility in soybean. B, C: RT-qPCR and RNA-seq analysis of nuclear genes related to cytoplasmic male sterility in soybean. D: RT-qPCR analysis of PPR genes. *, **, *** indicate significant difference at P<0.05, P<0.01, P<0.001 levels, respectively
| [1] | 刘杰, 黄学辉. 作物杂种优势研究现状与展望[J]. 中国科学: 生命科学, 2021, 51(10): 1396-1404. |
| Liu J, Huang XH. Advances and perspectives in crop heterosis[J]. Sci Sin Vitae, 2021, 51(10): 1396-1404. | |
| [2] |
Tester M, Langridge P. Breeding technologies to increase crop production in a changing world[J]. Science, 2010, 327(5967): 818-822.
doi: 10.1126/science.1183700 pmid: 20150489 |
| [3] | 王茜, 陈景斌, 林云, 等. 豆类作物雄性不育及杂种优势利用的研究进展[J]. 江苏农业科学, 2022, 50(4): 9-16. |
| Wang Q, Chen JB, Lin Y, et al. Research progress on male sterility and heterosis utilization in legume crops[J]. Jiangsu Agric Sci, 2022, 50(4): 9-16. | |
| [4] | 孙妍妍, 赵丽梅, 张伟, 等. 大豆杂种优势利用研究进展[J]. 大豆科技, 2021(6): 26-35. |
| Sun YY, Zhao LM, Zhang W, et al. Research progress on utilization of soybean heterosis[J]. Soybean Sci Technol, 2021(6): 26-35. | |
| [5] | 杨景华, 张明方. 线粒体反向调控介导高等植物细胞质雄性不育发生机制[J]. 遗传, 2007, 29(10): 1173-1181. |
| Yang JH, Zhang MF. Mechanism of cytoplasmic male-sterility modulated by mitochondrial retrograde regulation in higher plants[J]. Hereditas, 2007, 29(10): 1173-1181. | |
| [6] | Ji JJ, Huang W, Yin CC, et al. Mitochondrial cytochrome c oxidase and F1Fo-ATPase dysfunction in peppers(Capsicum annuum L.) with cytoplasmic male sterility and its association with orf507 and ψatp6-2 genes[J]. Int J Mol Sci, 2013, 14(1): 1050-1068. |
| [7] | Hu J, Wang K, Huang WC, et al. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162[J]. Plant Cell, 2012, 24(1): 109-122. |
| [8] |
Luo DP, Xu H, Liu ZL, et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice[J]. Nat Genet, 2013, 45: 573-577.
doi: 10.1038/ng.2570 pmid: 23502780 |
| [9] |
Wang K, Gao F, Ji YX, et al. ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice[J]. New Phytol, 2013, 198(2): 408-418.
doi: 10.1111/nph.12180 pmid: 23437825 |
| [10] |
Chen LT, Liu YG. Male sterility and fertility restoration in crops[J]. Annu Rev Plant Biol, 2014, 65: 579-606.
doi: 10.1146/annurev-arplant-050213-040119 pmid: 24313845 |
| [11] | Xiao SL, Zang J, Pei YR, et al. Activation of mitochondrial orf355 gene expression by a nuclear-encoded DREB transcription factor causes cytoplasmic male sterility in maize[J]. Mol Plant, 2020, 13(9): 1270-1283. |
| [12] | He TT, Ding XL, Zhang H, et al. Comparative analysis of mitochondrial genomes of soybean cytoplasmic male-sterile lines and their maintainer lines[J]. Funct Integr Genomics, 2021, 21(1): 43-57. |
| [13] | Li JJ, Han SH, Ding XL, et al. Comparative transcriptome analysis between the cytoplasmic male sterile line NJCMS1A and its maintainer NJCMS1B in soybean(Glycine max(L.)Merr.)[J]. PLoS One, 2015, 10(5): e0126771. |
| [14] | Bai ZY, Ding XL, Zhang RJ, et al. Transcriptome analysis reveals the genes related to pollen abortion in a cytoplasmic male-sterile soybean(Glycine max(L.)Merr.)[J]. Int J Mol Sci, 2022, 23(20): 12227. |
| [15] | Zhang CB, Fu FY, Lin CJ, et al. MicroRNAs involved in regulatory cytoplasmic male sterility by analysis RNA-seq and small RNA-seq in soybean[J]. Front Genet, 2021, 12: 654146. |
| [16] | 燕厚兴. 大豆RN型细胞质雄性不育相关基因挖掘与初步鉴定[D]. 长春: 长春师范大学, 2022. |
| Yan HX. Exploration and preliminary identification of genes related to RN-type cytoplasmic male sterility in soybean[D]. Changchun: Changchun Normal University, 2022. | |
| [17] |
白志元, 杨玉花, 张瑞军. 不同恢复型大豆细胞质雄性不育杂种F1的转录组分析[J]. 植物遗传资源学报, 2022, 23(6): 1847-1855.
doi: 10.13430/j.cnki.jpgr.20220510001 |
| Bai ZY, Yang YH, Zhang RJ. Transcriptomic analysis of soybean cytoplasmic male sterile F1hybrids from pollination with different restorer types[J]. J Plant Genet Resour, 2022, 23(6): 1847-1855. | |
| [18] |
Zhao Z, Ding Z, Huang JJ, et al. Copy number variation of the restorer Rf4 underlies human selection of three-line hybrid rice breeding[J]. Nat Commun, 2023, 14: 7333.
doi: 10.1038/s41467-023-43009-4 pmid: 37957162 |
| [19] | Lin YN, Yang HL, Liu HM, et al. A P-type pentatricopeptide repeat protein ZmRF5 promotes 5' region partial cleavages of atp6c transcripts to restore the fertility of CMS-C maize by recruiting a splicing factor[J]. Plant Biotechnol J, 2023, 11:38073308. |
| [20] | 张井勇, 孙寰, 赵丽梅, 等. 大豆RN不育胞质不育与恢复类型的研究[J]. 大豆科学, 2010, 29(4): 559-564. |
| Zhang JY, Sun H, Zhao LM, et al. Classification of male-sterile lines with RN sterile cytoplasm and their restorers[J]. Soybean Sci, 2010, 29(4): 559-564. | |
| [21] | 白志元, 杨玉花, 张瑞军. 苗期短光照对三系杂交大豆及亲本育性的影响[J]. 中国农业大学学报, 2021, 26(9): 11-17. |
| Bai ZY, Yang YH, Zhang RJ. Effects of short light at seedling stage on fertility of three-line hybrid soybean and its parents[J]. J China Agric Univ, 2021, 26(9): 11-17. | |
| [22] | 门中华, 李生秀. 植物生物节律性研究进展[J]. 生物学杂志, 2009, 26(5): 53-55. |
| Men ZH, Li SX. Progress in the study of plant biorhythm[J]. J Biol, 2009, 26(5): 53-55. | |
| [23] |
Harmer SL. The circadian system in higher plants[J]. Annu Rev Plant Biol, 2009, 60: 357-377.
doi: 10.1146/annurev.arplant.043008.092054 pmid: 19575587 |
| [24] |
Ye JL, Duan Y, Hu G, et al. Identification of candidate genes and biosynthesis pathways related to fertility conversion by wheat KTM3315A transcriptome profiling[J]. Front plant sci, 2017, 8: 449.
doi: 10.3389/fpls.2017.00449 pmid: 28428792 |
| [25] | Yu JP, Zhao GL, Li W, et al. A single nucleotide polymorphism in an R2R3 MYB transcription factor gene triggers the male sterility in soybean ms6(Ames1)[J]. Theor Appl Genet, 2021, 134(11): 3661-3674. |
| [26] | 聂智星. 大豆新雄性不育细胞质的发掘, 核不育基因的定位和产量杂种优势的QTL-等位变异解析[D]. 南京: 南京农业大学, 2017. |
| Nie ZX. Discovery of a new cytoplasmic male-sterile source, detection of a nuclear male-sterile gene and QTL-allele constitutions of heterosis in soybean[Glycine max(L.)merr.][D]. Nanjing: Nanjing Agricultural University, 2017. | |
| [27] |
Uyttewaal M, Arnal N, Quadrado M, et al. Characterization of raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for ogura cytoplasmic male sterility[J]. Plant Cell, 2008, 20(12): 3331-3345.
doi: 10.1105/tpc.107.057208 pmid: 19098270 |
| [28] | Yang SP, Duan MP, Meng QC, et al. Inheritance and gene tagging of male fertility restoration of cytoplasmic-nuclear male-sterile line NJCMS1A in soybean[J]. Plant Breed, 2007, 126(3): 302-305. |
| [29] | Wang TL, He TT, Ding XL, et al. Confirmation of GmPPR576 as a fertility restorer gene of cytoplasmic male sterility in soybean[J]. J Exp Bot, 2021, 72(22): 7729-7742. |
| [30] | Sun YY, Zhang Y, Jia SG, et al. Identification of a candidate restorer-of-fertility gene Rf3 encoding a pentatricopeptide repeat protein for the cytoplasmic male sterility in soybean[J]. Int J Mol Sci, 2022, 23: 5388. |
| [1] | ZHANG Zhen, LI Qing, XU Jing, CHEN Kai-yuan, ZHANG Chun-zhi, ZHU Guang-tao. Construction and Application of Potato Mitochondrial Targeted Expression Vector [J]. Biotechnology Bulletin, 2024, 40(5): 66-73. |
| [2] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [3] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [4] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [5] | LI Wen-chen, LIU Xin, KANG Yue, LI Wei, QI Ze-zheng, YU Lu, WANG Fang. Optimization and Application of Tobacco Rattle Virus-induced Gene Silencing System in Soybean [J]. Biotechnology Bulletin, 2023, 39(7): 143-150. |
| [6] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [7] | HOU Xiao-yuan, CHE Zheng-zheng, LI Heng-jing, DU Chong-yu, XU Qian, WANG Qun-qing. Construction of the Soybean Membrane System cDNA Library and Interaction Proteins Screening for Effector PsAvr3a [J]. Biotechnology Bulletin, 2023, 39(4): 268-276. |
| [8] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [9] | WANG Ge-ge, QIU Shi-rui, ZHANG Lin-han, YANG Guo-wei, XU Xiao-yun, WANG Ai-ling, ZENG Shu-hua, LIU Ya-jie. Molecular Cytology at Meiosis in Allotriploid Nicotiana tabacum(SST) [J]. Biotechnology Bulletin, 2023, 39(2): 183-192. |
| [10] | CHEN Yi-bo, YANG Wan-ming, YUE Ai-qin, WANG Li-xiang, DU Wei-jun, WANG Min. Construction of Soybean Genetic Map Based on SLAF Markers and QTL Mapping Analysis of Salt Tolerance at Seedling Stage [J]. Biotechnology Bulletin, 2023, 39(2): 70-79. |
| [11] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [12] | BAI Miao, TIAN Wen-qing, WU Shuai, WANG Min, WANG Li-xiang, YUE Ai-qin, NIU Jing-ping, ZHANG Yong-po, GAO Chun-yan, ZHANG Wu-xia, GUO Shu-jin, DU Wei-jun, ZHAO Jin-zhong. Effects of Hormonal and Adversely Stress on Vitamin E and γ-TMT Gene Expression in Soybeans [J]. Biotechnology Bulletin, 2023, 39(10): 148-162. |
| [13] | YU Hui-lin, WU Kong-ming. Commercialization Strategy of Transgenic Soybean in China [J]. Biotechnology Bulletin, 2023, 39(1): 1-15. |
| [14] | GUO Bin-hui, SONG Li. Transcription of Ethylene Biosynthesis and Signaling Associated Genes in Response to Heterodera glycine Infection [J]. Biotechnology Bulletin, 2022, 38(8): 150-158. |
| [15] | SHI Guang-cheng, YANG Wan-ming, DU Wei-jun, WANG Min. Screening of Salt-tolerant Soybean Germplasm and Physiological Characteristics Analysis of Its Salt Tolerance [J]. Biotechnology Bulletin, 2022, 38(4): 174-183. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||