








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 150-162.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1090
Previous Articles Next Articles
ZHANG Ming-ya( ), PANG Sheng-qun(
), PANG Sheng-qun( ), LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong
), LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong
Received:2023-11-20
Online:2024-07-26
Published:2024-07-30
Contact:
PANG Sheng-qun
E-mail:2419538996@qq.com;pangshqok@shzu.edu.cn
ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum[J]. Biotechnology Bulletin, 2024, 40(7): 150-162.
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| Actin | ATCTTGGCTTCCCTCAGCAC | GCATCTCTGGTCCAGTAGGAAAT |
| SlFAD1 | TCTACGGTGTGCCCCTTCTA | AGTGATGGGTGAGTGTGCTG |
| SlFAD4 | TCAAGAAAGCCATCCCACCC | TGGCAGATCCAGTAAAGCGG |
| SlFAD6 | CTCTTATCACCACCGCCCTC | GCTGCATTCACGATTGGCAT |
| SlFAD14 | CTTCCGTTTCTGTTTCCGCC | TAAGCGGTGCACTTACCCTC |
| SlFAD18 | CACCTGGTCTGTGGGAGTTC | CCAGCCCCAGAAGTAAACCA |
| SlFAD21 | CGAGGTTGAGGTGCTTGACT | TTCCAGGTTGAGTCGGATGC |
| SlFAD23 | CCGTTACTTTTGCATGGCCT | CCCAAGCTAGCCCTTTCATC |
Table 1 Primers for RT-qPCR
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| Actin | ATCTTGGCTTCCCTCAGCAC | GCATCTCTGGTCCAGTAGGAAAT |
| SlFAD1 | TCTACGGTGTGCCCCTTCTA | AGTGATGGGTGAGTGTGCTG |
| SlFAD4 | TCAAGAAAGCCATCCCACCC | TGGCAGATCCAGTAAAGCGG |
| SlFAD6 | CTCTTATCACCACCGCCCTC | GCTGCATTCACGATTGGCAT |
| SlFAD14 | CTTCCGTTTCTGTTTCCGCC | TAAGCGGTGCACTTACCCTC |
| SlFAD18 | CACCTGGTCTGTGGGAGTTC | CCAGCCCCAGAAGTAAACCA |
| SlFAD21 | CGAGGTTGAGGTGCTTGACT | TTCCAGGTTGAGTCGGATGC |
| SlFAD23 | CCGTTACTTTTGCATGGCCT | CCCAAGCTAGCCCTTTCATC |
| 保守基序名称 Conservative motif name | 具体序列分布情况 Specific sequence distribution |
|---|---|
| Motif 1 | WVTAHECGHHAFSDYQWJBDTVGFILHSSJLVPYFSWKYSHRRHHSNTGS |
| Motif 2 | NKVFHNITDTHVLHHLFSTIPHYHAVEATKAIKPLLGEYYQFDGTPIYKA |
| Motif 3 | EVKKHNKAKDCWJIISGKVYDVTKFLDDHPGGDEVLLSATG |
| Motif 4 | RVPSSKPPFTJSDIKKAIPPHCFQRSLVRSFSYVVRDJVLVFILYYIAAT |
| Motif 5 | CIYGVPLLIVNGFIVLITYLHHTHASLPHYDSSEWDWLRGA |
| Motif 6 | NVSGRPYDRFASHYBPYSPJYNDRERLQI |
| Motif 7 | KDATDDFEDVGHSSSAREMLDKYYIGEIDSSTIPTKRKYTP |
| Motif 8 | IWRDFKECIYVEKDEESQDKGVFWYKNK |
| Motif 9 | PYNYLAWPIYWIAQGCVFTGI |
| Motif 10 | QPKGNDWFEKQTAGTIDIACSPQMDWFFGGLQFQLEHHLFPRLPRCQLRK |
Table 2 Distribution of conservative motif sequence
| 保守基序名称 Conservative motif name | 具体序列分布情况 Specific sequence distribution |
|---|---|
| Motif 1 | WVTAHECGHHAFSDYQWJBDTVGFILHSSJLVPYFSWKYSHRRHHSNTGS |
| Motif 2 | NKVFHNITDTHVLHHLFSTIPHYHAVEATKAIKPLLGEYYQFDGTPIYKA |
| Motif 3 | EVKKHNKAKDCWJIISGKVYDVTKFLDDHPGGDEVLLSATG |
| Motif 4 | RVPSSKPPFTJSDIKKAIPPHCFQRSLVRSFSYVVRDJVLVFILYYIAAT |
| Motif 5 | CIYGVPLLIVNGFIVLITYLHHTHASLPHYDSSEWDWLRGA |
| Motif 6 | NVSGRPYDRFASHYBPYSPJYNDRERLQI |
| Motif 7 | KDATDDFEDVGHSSSAREMLDKYYIGEIDSSTIPTKRKYTP |
| Motif 8 | IWRDFKECIYVEKDEESQDKGVFWYKNK |
| Motif 9 | PYNYLAWPIYWIAQGCVFTGI |
| Motif 10 | QPKGNDWFEKQTAGTIDIACSPQMDWFFGGLQFQLEHHLFPRLPRCQLRK |
| 基因名称Gene name | 脱落酸响应Abscisic acid response | 厌氧诱导响应Anaerobic induction | 生长素响应Auxin response | 昼夜节律调控Circadian control | 防御和应激响应 Defense and stress response | 干旱诱导响应Drought- inducibility | 低温响应Low-temperature response | 赤霉素响应Gibberellin- response | 分生组织表达Meristem expression | 水杨酸响应Salicylic acid response | 茉莉酸响应MeJA- response |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SlFAD1 | 5 | 4 | 0 | 0 | 1 | 1 | 2 | 0 | 1 | 2 | 8 |
| SlFAD2 | 0 | 5 | 1 | 0 | 1 | 2 | 1 | 0 | 1 | 0 | 6 |
| SlFAD3 | 4 | 1 | 0 | 1 | 1 | 1 | 4 | 2 | 0 | 1 | 8 |
| SlFAD4 | 8 | 3 | 2 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 6 |
| SlFAD5 | 13 | 3 | 2 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 6 |
| SlFAD6 | 1 | 3 | 2 | 1 | 1 | 4 | 0 | 2 | 1 | 2 | 8 |
| SlFAD7 | 9 | 5 | 2 | 0 | 1 | 2 | 1 | 3 | 2 | 2 | 10 |
| SlFAD8 | 8 | 4 | 2 | 0 | 0 | 2 | 3 | 2 | 0 | 2 | 6 |
| SlFAD9 | 4 | 2 | 0 | 0 | 1 | 1 | 1 | 2 | 1 | 2 | 6 |
| SlFAD10 | 4 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 4 |
| SlFAD11 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 3 | 4 |
| SlFAD12 | 3 | 1 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 3 | 6 |
| SlFAD13 | 8 | 2 | 1 | 1 | 0 | 1 | 3 | 3 | 1 | 0 | 0 |
| SlFAD14 | 5 | 3 | 1 | 0 | 2 | 5 | 2 | 4 | 1 | 0 | 4 |
| SlFAD15 | 4 | 2 | 3 | 0 | 1 | 1 | 0 | 1 | 0 | 2 | 8 |
| SlFAD16 | 11 | 2 | 2 | 0 | 0 | 0 | 1 | 2 | 1 | 4 | 16 |
| SlFAD17 | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| SlFAD18 | 1 | 6 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 6 |
| SlFAD19 | 14 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 4 | 0 | 24 |
| SlFAD20 | 10 | 7 | 1 | 1 | 2 | 0 | 3 | 2 | 0 | 0 | 2 |
| SlFAD21 | 5 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 6 |
| SlFAD22 | 5 | 1 | 0 | 3 | 1 | 0 | 1 | 3 | 2 | 2 | 6 |
| SlFAD23 | 4 | 3 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 2 | 10 |
| SlFAD24 | 3 | 6 | 0 | 3 | 2 | 2 | 3 | 0 | 0 | 2 | 8 |
| SlFAD25 | 1 | 4 | 2 | 1 | 2 | 1 | 0 | 2 | 2 | 1 | 4 |
| SlFAD26 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 2 | 8 |
Table 3 Analysis of promoter cis-acting elements of the tomato SlFAD gene family
| 基因名称Gene name | 脱落酸响应Abscisic acid response | 厌氧诱导响应Anaerobic induction | 生长素响应Auxin response | 昼夜节律调控Circadian control | 防御和应激响应 Defense and stress response | 干旱诱导响应Drought- inducibility | 低温响应Low-temperature response | 赤霉素响应Gibberellin- response | 分生组织表达Meristem expression | 水杨酸响应Salicylic acid response | 茉莉酸响应MeJA- response |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SlFAD1 | 5 | 4 | 0 | 0 | 1 | 1 | 2 | 0 | 1 | 2 | 8 |
| SlFAD2 | 0 | 5 | 1 | 0 | 1 | 2 | 1 | 0 | 1 | 0 | 6 |
| SlFAD3 | 4 | 1 | 0 | 1 | 1 | 1 | 4 | 2 | 0 | 1 | 8 |
| SlFAD4 | 8 | 3 | 2 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 6 |
| SlFAD5 | 13 | 3 | 2 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 6 |
| SlFAD6 | 1 | 3 | 2 | 1 | 1 | 4 | 0 | 2 | 1 | 2 | 8 |
| SlFAD7 | 9 | 5 | 2 | 0 | 1 | 2 | 1 | 3 | 2 | 2 | 10 |
| SlFAD8 | 8 | 4 | 2 | 0 | 0 | 2 | 3 | 2 | 0 | 2 | 6 |
| SlFAD9 | 4 | 2 | 0 | 0 | 1 | 1 | 1 | 2 | 1 | 2 | 6 |
| SlFAD10 | 4 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 4 |
| SlFAD11 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 3 | 4 |
| SlFAD12 | 3 | 1 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 3 | 6 |
| SlFAD13 | 8 | 2 | 1 | 1 | 0 | 1 | 3 | 3 | 1 | 0 | 0 |
| SlFAD14 | 5 | 3 | 1 | 0 | 2 | 5 | 2 | 4 | 1 | 0 | 4 |
| SlFAD15 | 4 | 2 | 3 | 0 | 1 | 1 | 0 | 1 | 0 | 2 | 8 |
| SlFAD16 | 11 | 2 | 2 | 0 | 0 | 0 | 1 | 2 | 1 | 4 | 16 |
| SlFAD17 | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| SlFAD18 | 1 | 6 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 6 |
| SlFAD19 | 14 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 4 | 0 | 24 |
| SlFAD20 | 10 | 7 | 1 | 1 | 2 | 0 | 3 | 2 | 0 | 0 | 2 |
| SlFAD21 | 5 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 6 |
| SlFAD22 | 5 | 1 | 0 | 3 | 1 | 0 | 1 | 3 | 2 | 2 | 6 |
| SlFAD23 | 4 | 3 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 2 | 10 |
| SlFAD24 | 3 | 6 | 0 | 3 | 2 | 2 | 3 | 0 | 0 | 2 | 8 |
| SlFAD25 | 1 | 4 | 2 | 1 | 2 | 1 | 0 | 2 | 2 | 1 | 4 |
| SlFAD26 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 2 | 8 |
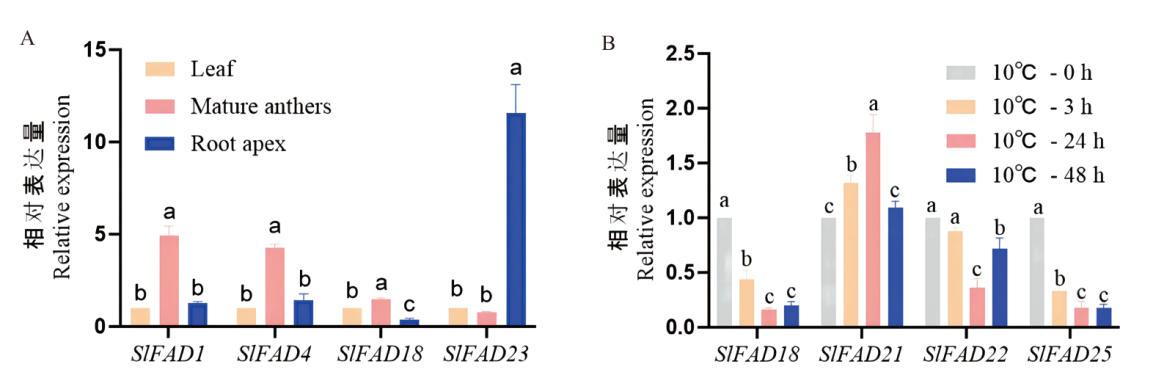
Fig. 6 Analysis of expression patterns of tomato SlFAD gene family members in different tissues and at 10℃ Different letters indicate significant differences between treatments at the 0.05 level
| [1] |
Li NN, Xu CC, Li-Beisson Y, et al. Fatty acid and lipid transport in plant cells[J]. Trends Plant Sci, 2016, 21(2): 145-158.
doi: S1360-1385(15)00259-9 pmid: 26616197 |
| [2] | Cheng CZ, Liu F, Sun XL, et al. Genome-wide identification of FAD gene family and their contributions to the temperature stresses and mutualistic and parasitic fungi colonization responses in banana[J]. Int J Biol Macromol, 2022, 204: 661-676. |
| [3] | 李凤, 郑汪东, 唐文博. 谷子膜结合脂肪酸脱氢酶基因家族的鉴定及进化分析[J]. 山西农业科学, 2019, 47(7): 1121-1125. |
| Li F, Zheng WD, Tang WB. Identification and evolutionary analysis of membrane-bound fatty acid desaturases gene family in Setaria italica[J]. J Shanxi Agric Sci, 2019, 47(7): 1121-1125. | |
| [4] | 崔景焱. 亚麻芥中脂肪酸脱氢酶基因克隆分析与功能验证[D]. 长春: 吉林农业大学, 2019. |
| Cui JY. Cloning and functional characterization of fatty acid dehydrogenase gene in Camelina sativa[D]. Changchun: Jilin Agricultural University, 2019. | |
| [5] | 朱宗文, 张爱冬, 吴雪霞, 等. 生物信息学鉴定分析茄子脂肪酸去饱和酶(FAD)基因家族[J]. 分子植物育种, 2023, 21(8): 2453-2463. |
| Zhu ZW, Zhang AD, Wu XX, et al. Identification and bioinformatics analysis of fatty acid desaturase(FAD)gene family in eggplant(Solanum melongena L.)[J]. Mol Plant Breed, 2023, 21(8): 2453-2463. | |
| [6] |
焦苏淇, 周俊名, 尚雨晴, 等. 大豆脂肪酸脱氢酶GmFAD3C-1基因的克隆及功能分析[J]. 中国油料作物学报, 2022, 44(5): 1006-1017.
doi: 10.19802/j.issn.1007-9084.2021253 |
| Jiao SQ, Zhou JM, Shang YQ, et al. Cloning and genetic transformation of soybean fatty acid dehydrogenase GmFAD3C-1 gene[J]. Chin J Oil Crop Sci, 2022, 44(5): 1006-1017. | |
| [7] | 侯静静, 赵利, 王斌. 亚麻FAD基因家族的生物信息学鉴定分析[J]. 寒旱农业科学, 2023, 2(3): 246-253. |
| Hou JJ, Zhao L, Wang B. Identification and bioinformatics analysis of FAD gene family in Linum usitatissimum L[J]. J Cold Arid Agric Sci, 2023, 2(3): 246-253. | |
| [8] | 于海彦, 周志峰, 贾俊强, 等. 家蚕类ω3-脂肪酸脱氢酶基因的克隆、表达及功能研究[J]. 蚕业科学, 2017, 43(4): 577-586. |
| Yu HY, Zhou ZF, Jia JQ, et al. Cloning, expression and functional analysis of an ω3-fatty acid desaturase-like gene from Bombyx mori[J]. Sci Seric, 2017, 43(4): 577-586. | |
| [9] | 叶浩盈. 三角褐指藻脂肪酸去饱和酶基因家族分析及高产EPA藻株的选育[D]. 福州: 福建师范大学, 2019. |
| Ye HY. Analysis of fatty acid desaturase gene family of Phaeodactylum tricornutum and breeding of high-yield EPA algae strain[D]. Fuzhou: Fujian Normal University, 2019. | |
| [10] | 刘永红. 白沙蒿△12-脂肪酸脱氢酶基因克隆及其植物表达载体构建[D]. 兰州: 兰州大学, 2010. |
| Liu YH. Cloning and expression vector construction of △12-fatty acid desaturase(FAD2) gene from Artemisia sphaero-cephala[D]. Lanzhou: Lanzhou University, 2010. | |
| [11] |
Thiede MA, Ozols J, Strittmatter P. Construction and sequence of cDNA for rat liver stearyl coenzyme A desaturase[J]. J Biol Chem, 1986, 261(28): 13230-13235.
pmid: 2428815 |
| [12] | 缪秀梅. 白沙蒿FAD2基因家族克隆与功能研究及转基因苜蓿评价[D]. 兰州: 兰州大学, 2020. |
| Miao XM. Cloning and functional analysis of FAD2 gene family of Artemisia sphaerocephala and evaluation of transgenic alfalfa[D]. Lanzhou: Lanzhou University, 2020. | |
| [13] |
薛晓梦, 李建国, 白冬梅, 等. 花生FAD2基因家族表达分析及其对低温胁迫的响应[J]. 作物学报, 2019, 45(10): 1586-1594.
doi: 10.3724/SP.J.1006.2019.84177 |
| Xue XM, Li JG, Bai DM, et al. Expression profiles of FAD2 genes and their responses to cold stress in peanut[J]. Acta Agron Sin, 2019, 45(10): 1586-1594. | |
| [14] | Lou Y, Schwender J, Shanklin J. FAD2 and FAD3 desaturases form heterodimers that facilitate metabolic channeling in vivo[J]. J Biol Chem, 2014, 289(26): 17996-18007. |
| [15] | 李丹丹, 王庆, 胡博, 等. 低温对红花脂肪酸组成和脂肪酸脱氢酶基因表达的影响[J]. 分子植物育种, 2018, 16(16): 5223-5231. |
| Li DD, Wang Q, Hu B, et al. Effects of low temperature on fatty acid composition and gene expression of fatty acid desaturase in safflower[J]. Mol Plant Breed, 2018, 16(16): 5223-5231. | |
| [16] |
常培培, 张静, 杨建华, 等. 紫色番茄果实挥发性风味物质分析[J]. 食品科学, 2014, 35(14): 165-169.
doi: 10.7506/spkx1002-6630-201414032 |
| Chang PP, Zhang J, Yang JH, et al. Analysis of volatile compounds in purple tomato fruits[J]. Food Sci, 2014, 35(14): 165-169. | |
| [17] | 宋慧慧, 颜龙飞, 苟莎莎, 等. PuFAD8基因过表达载体的构建及其在番茄中的遗传转化[J]. 合肥工业大学学报: 自然科学版, 2023, 46(3): 392-396. |
| Song HH, Yan LF, Gou SS, et al. Construction of overexpression vector of PuFAD8 gene and its genetic transformation in tomato(Solanum lycopersicum)[J]. J Hefei Univ Technol Nat Sci, 2023, 46(3): 392-396. | |
| [18] | Wang JJ, Liu HR, Gao J, et al. Two ω-3 FADs are associated with peach fruit volatile formation[J]. International journal of molecular sciences, 2016, 17(4): 464. |
| [19] | 马银银, 李莉娜, 刘琦, 等. 苹果香气物质的组成、影响因素及调控策略[J]. 北方园艺, 2023(1): 119-127. |
| Ma YY, Li LN, Liu Q, et al. Composition, influencing factors and regulation strategies of aroma substances in apple[J]. North Hortic, 2023(1): 119-127. | |
| [20] | 赵训超, 魏玉磊, 丁冬, 等. 甜荞麦脂肪酸脱氢酶基因(FeFAD)家族的鉴定与分析[J]. 东北农业科学, 2021, 46(1): 36-41. |
| Zhao XC, Wei YL, Ding D, et al. Genome-wide identification and bioinformatics analysis of fatty acid desaturase gene(FeFAD)family in common buckwheat[J]. J Northeast Agric Sci, 2021, 46(1): 36-41. | |
| [21] | 向妮艳. 红花膜结合脂肪酸脱氢酶基因家族的鉴定及表达分析[D]. 武汉: 中南民族大学, 2020. |
| Xiang NY. Identification and expression analysis of membrane-bound fatty acid desaturases gene family in safflower[D]. Wuhan: South-central University for Nationalities, 2020. | |
| [22] | Barrero-Gil J, Huertas R, Rambla JL, et al. Tomato plants increase their tolerance to low temperature in a chilling acclimation process entailing comprehensive transcriptional and metabolic adjustments[J]. Plant Cell Environ, 2016, 39(10): 2303-2318. |
| [23] | Penin AA, Klepikova AV, Kasianov AS, et al. Comparative analysis of developmental transcriptome maps of Arabidopsis thaliana and Solanum lycopersicum[J]. Genes, 2019, 10(1): 50. |
| [24] | 包鹏. 续随子ω-3脂肪酸脱氢酶基因ElFAD3的克隆和功能研究[D]. 太谷: 山西农业大学, 2022. |
| Bao P. Cloning and functional analysis of ω-3 fatty acid dehydrogenase ElFAD3 gene in Euphorbia lathyris[D]. Taigu: Shanxi Agricultural University, 2022. | |
| [25] | 章妮, 王欢, 李绍峰, 等. 毛竹FAD基因家族鉴定与综合分析[J/OL]. 分子植物育种, 2023: 1-17. http://kns.cnki.net/kcms/detail/46.1068.S.20230309.1448.004.html. |
| Zhang N, Wang H, Li SF, et al. Identification and comprehensive analysis of FAD gene family of Bamboo[J/OL]. Molecular Plant Breeding, 2023:1-17. http://kns.cnki.net/kcms/detail/46.1068.S.20230309.1448.004.html. | |
| [26] | 冯吉妤. 陆地棉脂肪酸脱氢酶基因家族的全基因组鉴定和表达特征分析[D]. 杭州: 浙江大学, 2017. |
| Feng JY. Genome-wide identification of fatty acid desaturase gene family in Gossypium hirsutum L. and their expression patterns[D]. Hangzhou: Zhejiang University, 2017. | |
| [27] | Zhao XC, Wei JP, He L, et al. Identification of fatty acid desaturases in maize and their differential responses to low and high temperature[J]. Genes, 2019, 10(6): 445. |
| [28] | 阮建, 单雷, 李新国, 等. 花生FAD基因家族的全基因组鉴定与表达模式分析[J]. 山东农业科学, 2018, 50(6): 1-9. |
| Ruan J, Shan L, Li XG, et al. Genome-wide identification and expression pattern analysis of peanut FAD gene family[J]. Shandong Agric Sci, 2018, 50(6): 1-9. | |
| [29] | 陈义珍, 李浩, 傅明川, 等. 陆地棉膜结合脂肪酸去饱和酶基因家族的全基因组鉴定及表达分析[J]. 山东农业科学, 2023, 55(4): 11-23. |
| Chen YZ, Li H, Fu MC, et al. Genome-wide identification and expression analysis of membrane-bound fatty acid desaturase gene family in upland cotton(Gossypium hirsutum L.)[J]. Shandong Agric Sci, 2023, 55(4): 11-23. | |
| [30] | 金兄莲, 安进宝, 祁得胜, 等. 毛白杨FAD基因家族鉴定与表达模式分析[J/OL]. 分子植物育种 2022: 1-23. http://kns.cnki.net/kcms/detail/46.1068.S.20220825.1145.006.html. |
| Jin XL, An JB, Qi DS, et al. Identification and expression pattern analysis of FAD gene family in Populus tomentosa[J/OL]. Molecular Plant Breeding 2022: 1-23. http://kns.cnki.net/kcms/detail/46.1068.S.20220825.1145.006.html. |
| [1] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [2] | LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis [J]. Biotechnology Bulletin, 2024, 40(6): 190-202. |
| [3] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [4] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [5] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [6] | HU Yong-bo, LEI Yu-tian, YANG Yong-sen, CHEN Xin, LIN Huang-fang, LIN Bi-ying, LIU Shuang, BI Ge, SHEN Bao-ying. Genome-wide Identification and Expression Pattern Analysis of the Bcl-2-related Anti-apoptotic Family in Cucumis sativus L. and Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2024, 40(6): 219-237. |
| [7] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [8] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [9] | HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit [J]. Biotechnology Bulletin, 2024, 40(5): 167-178. |
| [10] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [11] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [12] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [13] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [14] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [15] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||