








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (1): 333-346.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0575
Previous Articles Next Articles
RAO Jun1( ), ZHAO Chen1, LI Duan-hua1, LIAO Hao2, HUANG Jia-yu1, WANG Lu1(
), ZHAO Chen1, LI Duan-hua1, LIAO Hao2, HUANG Jia-yu1, WANG Lu1( )
)
Received:2024-06-14
Online:2025-01-26
Published:2025-01-22
Contact:
WANG Lu
E-mail:1274797431@qq.com;wanglu@cdu.edu.cn
RAO Jun, ZHAO Chen, LI Duan-hua, LIAO Hao, HUANG Jia-yu, WANG Lu. Application of Auto-induction Strategy in Ergothioneine Biosynthesis[J]. Biotechnology Bulletin, 2025, 41(1): 333-346.
| ERG合成途径来源 ERG synthesis pathway source | 关键酶* Key enzymes* | 工程菌 Engineering bacteria | 发酵工艺 Fermentation process | 诱导方式 Induction method | 发酵周期Fermentation period/h | 菌体密度 Bacterial density | ERG产量ERG production/(g·L-1) | ERG产率 ERG productivity/(g·L-1·h-1) | 参考文献 Reference |
|---|---|---|---|---|---|---|---|---|---|
| Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 73 | - | 0.6 | 8.2 | [ |
| Bacteria | W | Corynebacterium glutamicum | Fed-batch baffled flasks | - | 120 | OD600>80 | 0.1 | 0.8 | [ |
| Bacteria | W | E. coli | Baffled flasks | Manual addition of inducers | 192 | OD600=14.0 | 0.7 | 3.4 | [ |
| Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 216 | OD600≈55 | 1.3 | 6.1 | [ |
| Fungi | W | Yarrowia lipolytica | Fed-batch jar fermentation | - | 168 | OD600>150 | 7.3 | 43.5 | [ |
| Fungi | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 143 | OD600=130 | 4.3 | 30.3 | [ |
| Fungi | W | Yarrowia lipolytica | Fed-batch jar fermentation | - | 220 | 60.6 g/L CDW | 1.6 | 7.4 | [ |
| Fungi+Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 96 | OD600>75 | 2.6 | 27.4 | [ |
| Fungi+Bacteria | M | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 94 | OD600≈100 | 5.4 | 57.4 | [ |
| Fungi+Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 108 | OD600=45.7 | 0.7 | 6.6 | [ |
| Fungi+Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 108 | OD600=65 | 2 | 18.6 | [ |
Table 1 Fermentation levels for biosynthesis of ergothioneine by microbial liquid fermentation in recent years
| ERG合成途径来源 ERG synthesis pathway source | 关键酶* Key enzymes* | 工程菌 Engineering bacteria | 发酵工艺 Fermentation process | 诱导方式 Induction method | 发酵周期Fermentation period/h | 菌体密度 Bacterial density | ERG产量ERG production/(g·L-1) | ERG产率 ERG productivity/(g·L-1·h-1) | 参考文献 Reference |
|---|---|---|---|---|---|---|---|---|---|
| Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 73 | - | 0.6 | 8.2 | [ |
| Bacteria | W | Corynebacterium glutamicum | Fed-batch baffled flasks | - | 120 | OD600>80 | 0.1 | 0.8 | [ |
| Bacteria | W | E. coli | Baffled flasks | Manual addition of inducers | 192 | OD600=14.0 | 0.7 | 3.4 | [ |
| Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 216 | OD600≈55 | 1.3 | 6.1 | [ |
| Fungi | W | Yarrowia lipolytica | Fed-batch jar fermentation | - | 168 | OD600>150 | 7.3 | 43.5 | [ |
| Fungi | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 143 | OD600=130 | 4.3 | 30.3 | [ |
| Fungi | W | Yarrowia lipolytica | Fed-batch jar fermentation | - | 220 | 60.6 g/L CDW | 1.6 | 7.4 | [ |
| Fungi+Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 96 | OD600>75 | 2.6 | 27.4 | [ |
| Fungi+Bacteria | M | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 94 | OD600≈100 | 5.4 | 57.4 | [ |
| Fungi+Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 108 | OD600=45.7 | 0.7 | 6.6 | [ |
| Fungi+Bacteria | W | E. coli | Fed-batch jar fermentation | Manual addition of inducers | 108 | OD600=65 | 2 | 18.6 | [ |

Fig. 1 Metabolic pathways of ergothioneine and its precursor amino acids Introduction of the ERG synthesis gene(egtABCDE)from Mycobacterium smegmatis into E. coli results in ERG synthesis. Methionine, histidine, cysteine, and glutamate are the major precursors in this pathway, with glutamate being recycled and not physically consumed. HisG is the key enzyme in the synthesis of histidine whereas SerA and CysE are the key enzymes in the synthesis of cysteine. γGC: γ-glutamylcysteine; SAM: S-Adenosyl methionine; SAH: S-adenosyl-L-homocysteine
| 类型 Type | 名称 Name | 描述 Description | 来源 Source |
|---|---|---|---|
| Plasmid | pET-28a | Expression vector, Kanr | Lab stock |
| pET28a-egtABCDE | pET-28a containing egtABCDE | This work | |
| pET28a-egtA | pET-28a containing egtA | This work | |
| pET28a-egtB | pET-28a containing egtB | This work | |
| pET28a-egtC | pET-28a containing egtC | This work | |
| pET28a-egtD | pET-28a containing egtD | This work | |
| pET28a-egtE | pET-28a containing egtE | This work | |
| pACYC184 | Expression vector, Cmr, Tcr | General Biol | |
| pACYC184-cysE-serA | pACYC184 containing cysET167A, G203S, T234S, P252L, M256Q and serAT410stop | This work | |
| pACYC184-hisG | pACYC184 containing hisGS143F, ∆209-281 | This work | |
| pACYC184-hisG-cysE-serA | pACYC184 containing cysET167A, G203S, T234S, P252L, M256Q, serAT410stopand hisGS143F, ∆209-281 | This work | |
| Strain | E. coli Rosetta2(DE3) | Expression host | Lab stock |
| RE | E. coli Rosetta2(DE3)harboring pET28a-egtABCDE | This work | |
| REA | E. coli Rosetta2(DE3)harboring pET28a-egtA | This work | |
| REB | E. coli Rosetta2(DE3)harboring pET28a-egtB | This work | |
| REC | E. coli Rosetta2(DE3)harboring pET28a-egtC | This work | |
| RED | E. coli Rosetta2(DE3)harboring pET28a-egtD | This work | |
| REE | E. coli Rosetta2(DE3)harboring pET28a-egtE | This work | |
| RE-C | RE harboring pACYC184-cysE-serA | This work | |
| RE-H | RE harboring pACYC184-hisG | This work | |
| RE-CH | RE harboring pACYC184-hisG-cysE-serA | This work |
Table 2 Plasmids and strains
| 类型 Type | 名称 Name | 描述 Description | 来源 Source |
|---|---|---|---|
| Plasmid | pET-28a | Expression vector, Kanr | Lab stock |
| pET28a-egtABCDE | pET-28a containing egtABCDE | This work | |
| pET28a-egtA | pET-28a containing egtA | This work | |
| pET28a-egtB | pET-28a containing egtB | This work | |
| pET28a-egtC | pET-28a containing egtC | This work | |
| pET28a-egtD | pET-28a containing egtD | This work | |
| pET28a-egtE | pET-28a containing egtE | This work | |
| pACYC184 | Expression vector, Cmr, Tcr | General Biol | |
| pACYC184-cysE-serA | pACYC184 containing cysET167A, G203S, T234S, P252L, M256Q and serAT410stop | This work | |
| pACYC184-hisG | pACYC184 containing hisGS143F, ∆209-281 | This work | |
| pACYC184-hisG-cysE-serA | pACYC184 containing cysET167A, G203S, T234S, P252L, M256Q, serAT410stopand hisGS143F, ∆209-281 | This work | |
| Strain | E. coli Rosetta2(DE3) | Expression host | Lab stock |
| RE | E. coli Rosetta2(DE3)harboring pET28a-egtABCDE | This work | |
| REA | E. coli Rosetta2(DE3)harboring pET28a-egtA | This work | |
| REB | E. coli Rosetta2(DE3)harboring pET28a-egtB | This work | |
| REC | E. coli Rosetta2(DE3)harboring pET28a-egtC | This work | |
| RED | E. coli Rosetta2(DE3)harboring pET28a-egtD | This work | |
| REE | E. coli Rosetta2(DE3)harboring pET28a-egtE | This work | |
| RE-C | RE harboring pACYC184-cysE-serA | This work | |
| RE-H | RE harboring pACYC184-hisG | This work | |
| RE-CH | RE harboring pACYC184-hisG-cysE-serA | This work |
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| EgtA-F | ATATACCATGGCCCTGCCGG |
| EgtA-R | CCCAAGCTTTCCTTCTTACAGTTCACCTTTTGCC |
| EgtB-F | CATGCCATGGAACTGATTGCACGTGAAACCCTGG |
| EgtB-R | CCCAAGCTTCTCCTTCTTACACATCCCATGCC |
| EgtC-F | CATGCCATGGAATGTCGCCATGTTGCCTGGCT |
| EgtC-R | CCCAAGCTTCTTCTTACAGCGGGGTAACAAC |
| EgtD-F | CATGCCATGGAAACCCTGAGCCTGGCCAATTATC |
| EgtD-R | CCCAAGCTTCCTTCTTAACGAACTGCCAGGC |
| EgtE-F | CATGCCATGGAAATGCTGGCACAGCAGTGGCGTGA |
| EgtE-R | CCCAAGCTTAGCTTTTACGGTGCTTCACGC |
| HisG-F | ATAAAATATTTCTAGTTTTTTTCATATGCCTGACGGAGTTCACAC |
| HisG-R | TGCACTGAAATCTAGTTAATTCTGTGCATGCAGAATACCCT |
Table 3 Related primers
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| EgtA-F | ATATACCATGGCCCTGCCGG |
| EgtA-R | CCCAAGCTTTCCTTCTTACAGTTCACCTTTTGCC |
| EgtB-F | CATGCCATGGAACTGATTGCACGTGAAACCCTGG |
| EgtB-R | CCCAAGCTTCTCCTTCTTACACATCCCATGCC |
| EgtC-F | CATGCCATGGAATGTCGCCATGTTGCCTGGCT |
| EgtC-R | CCCAAGCTTCTTCTTACAGCGGGGTAACAAC |
| EgtD-F | CATGCCATGGAAACCCTGAGCCTGGCCAATTATC |
| EgtD-R | CCCAAGCTTCCTTCTTAACGAACTGCCAGGC |
| EgtE-F | CATGCCATGGAAATGCTGGCACAGCAGTGGCGTGA |
| EgtE-R | CCCAAGCTTAGCTTTTACGGTGCTTCACGC |
| HisG-F | ATAAAATATTTCTAGTTTTTTTCATATGCCTGACGGAGTTCACAC |
| HisG-R | TGCACTGAAATCTAGTTAATTCTGTGCATGCAGAATACCCT |
| 氨基酸 Amino acid | 终浓度Final concentration/(g·L-1) |
|---|---|
| Glu | 0.8 |
| 1.4 | |
| 2.0 | |
| His | 1.4 |
| 2.0 | |
| 2.6 | |
| Cys | 1.4 |
| 1.8 | |
| 2.0 | |
| Met | 0.25 |
| 0.5 | |
| 1.0 | |
| BLK | 0.0 |
Table 4 Added concentration of precursor amino acid
| 氨基酸 Amino acid | 终浓度Final concentration/(g·L-1) |
|---|---|
| Glu | 0.8 |
| 1.4 | |
| 2.0 | |
| His | 1.4 |
| 2.0 | |
| 2.6 | |
| Cys | 1.4 |
| 1.8 | |
| 2.0 | |
| Met | 0.25 |
| 0.5 | |
| 1.0 | |
| BLK | 0.0 |
| 水平 Level | 因素 Factors | ||
|---|---|---|---|
| A: Glu/(g·L-1) | B: His/(g·L-1) | C: Cys/(g·L-1) | |
| 1 | 1.1 | 1.7 | 1.6 |
| 2 | 1.4 | 2.0 | 1.8 |
| 3 | 1.7 | 2.3 | 2.0 |
Table 5 Factors and levels of orthogonal experiments
| 水平 Level | 因素 Factors | ||
|---|---|---|---|
| A: Glu/(g·L-1) | B: His/(g·L-1) | C: Cys/(g·L-1) | |
| 1 | 1.1 | 1.7 | 1.6 |
| 2 | 1.4 | 2.0 | 1.8 |
| 3 | 1.7 | 2.3 | 2.0 |
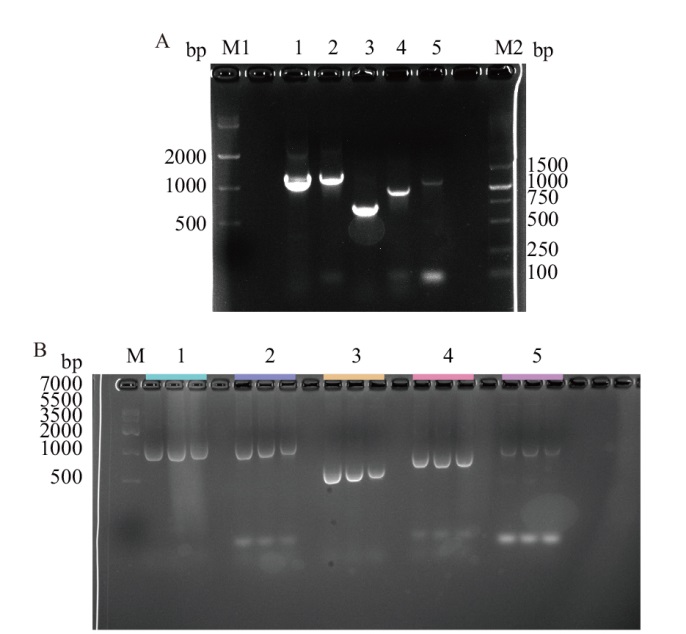
Fig. 2 Construction of positive engineering strain A: Amplified results of a single egt gene(M1:DNA marker IV;M2:DNA marker DL5000;1-5:egtA, egtB, egtC, egtD, and egtE)). B: Results of PCR identification(M:DNA marker IV;1-5:egtA,egtB,egtC, egtD, and egtE)
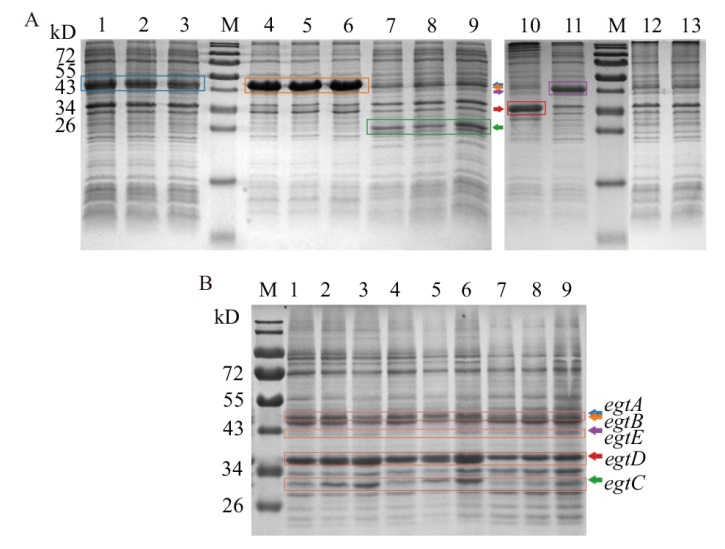
Fig. 3 Identification results of engineering bacteria expression A: SDS-PAGE results of single gene expression(M: Protein marker; 1-3: expression results for egtA; 4-6: expression results for egtB; 7-9: expression results for egtC; 10-12: expression results of egtD, egtE and NC(E. coli Rosetta2(DE3)/pET28a)respectively). B: RE strain expression SDS-PAGE results(M: Protein marker; 1-3, 4-6: expression results of RE strain at 2, 4, and 20 h after induction, respectively; 7-9: expression results of NC)

Fig. 4 Determination of ergothioneine content in liquid fermentation broth A: HPLC quantitation profile. B: LC-MS detection outcomes. i: The HPLC results of the supernatant of the fermentation broth(using an amino column)show that the retention time of the target compounds is 17.98 min; ii: m/z result plots for peaks with retention times in the range of(17.985 1±0.058 5)min. C: Comparison of HPLC results of fermentation products and ERG standards
| 诱导剂 Inducers | 诱导温度 Induction temperature/℃ | 麦角硫因含量 ERG content/(mg·L-1) | 上清中蛋白含量 Protein content of supernatant | 沉淀中蛋白含量 Protein content of precipitation |
|---|---|---|---|---|
| 0.4 mmol/L IPTG | 37 | 10.7 | + | +++ |
| 28 | 17.3 | ++ | ++ | |
| 16 | 9.8 | +++ | + | |
| 0.8% lactose | 37 | 5.4 | + | ++ |
| 28 | 10.2 | + | + | |
| 16 | 3.3 | + | + |
Table 6 Optimization of expression conditions
| 诱导剂 Inducers | 诱导温度 Induction temperature/℃ | 麦角硫因含量 ERG content/(mg·L-1) | 上清中蛋白含量 Protein content of supernatant | 沉淀中蛋白含量 Protein content of precipitation |
|---|---|---|---|---|
| 0.4 mmol/L IPTG | 37 | 10.7 | + | +++ |
| 28 | 17.3 | ++ | ++ | |
| 16 | 9.8 | +++ | + | |
| 0.8% lactose | 37 | 5.4 | + | ++ |
| 28 | 10.2 | + | + | |
| 16 | 3.3 | + | + |

Fig. 5 Effects of precursor amino acids on ERG production A: Results of one-factor experiments with exogenously added precursor amino acids. B: Chassis cell modification shake flask validation; *P≤0.05, **P≤0.01
| 实验号 Experiment No. | 列号Column number | ERG产量 ERG yield/(mg·L-1) | ||
|---|---|---|---|---|
| A | B | C | ||
| 1 | 1 | 1 | 1 | 77.4±4.2 |
| 2 | 1 | 2 | 3 | 81.8±1.4 |
| 3 | 1 | 3 | 2 | 81.4±2.0 |
| 4 | 2 | 1 | 3 | 82.8±1.9 |
| 5 | 2 | 2 | 2 | 89.7±2.0 |
| 6 | 2 | 3 | 1 | 82.5±1.2 |
| 7 | 3 | 1 | 2 | 80.8±2.2 |
| 8 | 3 | 2 | 1 | 82.7±3.5 |
| 9 | 3 | 3 | 3 | 91.2±6.0 |
| Ij | 240.7 | 241.0 | 242.7 | T=750.5 |
| IIj | 255.1 | 254.3 | 251.9 | |
| IIIj | 254.8 | 255.2 | 255.9 | |
| Rj | 14.4 | 14.2 | 13.2 | |
Table 7 Results of orthogonal experiments
| 实验号 Experiment No. | 列号Column number | ERG产量 ERG yield/(mg·L-1) | ||
|---|---|---|---|---|
| A | B | C | ||
| 1 | 1 | 1 | 1 | 77.4±4.2 |
| 2 | 1 | 2 | 3 | 81.8±1.4 |
| 3 | 1 | 3 | 2 | 81.4±2.0 |
| 4 | 2 | 1 | 3 | 82.8±1.9 |
| 5 | 2 | 2 | 2 | 89.7±2.0 |
| 6 | 2 | 3 | 1 | 82.5±1.2 |
| 7 | 3 | 1 | 2 | 80.8±2.2 |
| 8 | 3 | 2 | 1 | 82.7±3.5 |
| 9 | 3 | 3 | 3 | 91.2±6.0 |
| Ij | 240.7 | 241.0 | 242.7 | T=750.5 |
| IIj | 255.1 | 254.3 | 251.9 | |
| IIIj | 254.8 | 255.2 | 255.9 | |
| Rj | 14.4 | 14.2 | 13.2 | |
| 工艺 Process | 菌株 Strain | 补料 Feeding | 发酵规模 Scale/L | 周期 Period/h | 湿重Wet weight/(g·L-1) | 产量Yield/(g·L-1) | 产率Productivity/(mg·L-1·h-1) | 单位湿重菌体产量Yield per wet weight/(mg·g-1) |
|---|---|---|---|---|---|---|---|---|
| 常规诱导Routine induction | RE | Met | 10 | 142.0 | 12.3 | 0.2 | 1.2 | 13.7 |
| 自诱导 Self-induced | RE | Met | 10 | 135.5 | 123.2 | 0.4 | 3.1 | 3.4 |
| RE-CH | Met | 10 | 114.6 | 84.8 | 0.5 | 4.3 | 5.8 | |
| 自诱导优化Self-induced optimization | RE-CH | Met | 10 | 118.0 | 91.5 | 1.1 | 9.7 | 12.0 |
| 自诱导放大Self-induced scale-up | RE-CH | Met, His, Cys | 30 | 95.5 | 142.6 | 4.3 | 45.2 | 30.2 |
Table 8 Results of fermentation experiments in 10 L and 30 L bioreactor
| 工艺 Process | 菌株 Strain | 补料 Feeding | 发酵规模 Scale/L | 周期 Period/h | 湿重Wet weight/(g·L-1) | 产量Yield/(g·L-1) | 产率Productivity/(mg·L-1·h-1) | 单位湿重菌体产量Yield per wet weight/(mg·g-1) |
|---|---|---|---|---|---|---|---|---|
| 常规诱导Routine induction | RE | Met | 10 | 142.0 | 12.3 | 0.2 | 1.2 | 13.7 |
| 自诱导 Self-induced | RE | Met | 10 | 135.5 | 123.2 | 0.4 | 3.1 | 3.4 |
| RE-CH | Met | 10 | 114.6 | 84.8 | 0.5 | 4.3 | 5.8 | |
| 自诱导优化Self-induced optimization | RE-CH | Met | 10 | 118.0 | 91.5 | 1.1 | 9.7 | 12.0 |
| 自诱导放大Self-induced scale-up | RE-CH | Met, His, Cys | 30 | 95.5 | 142.6 | 4.3 | 45.2 | 30.2 |
| 发酵周期 Period/h | 上清Supernatant | 菌体Microorganism | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cys | Met | His | γGC | Cys | Met | His | γGC | ||
| 15.9 | - | + | - | - | - | + | - | - | |
| 20.7 | - | + | - | - | - | + | - | - | |
| 38.4 | - | - | - | - | - | + | + | + | |
| 43.8 | - | - | - | - | - | - | - | + | |
| 46.3 | - | - | - | - | - | + | + | + | |
| 62.2 | - | + | - | - | - | + | - | + | |
| 86.4 | - | + | - | - | - | + | - | + | |
| 95.0 | - | + | - | - | - | + | + | + | |
| 111.0 | - | + | - | - | - | + | + | + | |
| 114.6 | - | + | - | - | - | + | - | + | |
Table 9 Determination of major precursors and important intermediates in fermentation broths
| 发酵周期 Period/h | 上清Supernatant | 菌体Microorganism | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cys | Met | His | γGC | Cys | Met | His | γGC | ||
| 15.9 | - | + | - | - | - | + | - | - | |
| 20.7 | - | + | - | - | - | + | - | - | |
| 38.4 | - | - | - | - | - | + | + | + | |
| 43.8 | - | - | - | - | - | - | - | + | |
| 46.3 | - | - | - | - | - | + | + | + | |
| 62.2 | - | + | - | - | - | + | - | + | |
| 86.4 | - | + | - | - | - | + | - | + | |
| 95.0 | - | + | - | - | - | + | + | + | |
| 111.0 | - | + | - | - | - | + | + | + | |
| 114.6 | - | + | - | - | - | + | - | + | |
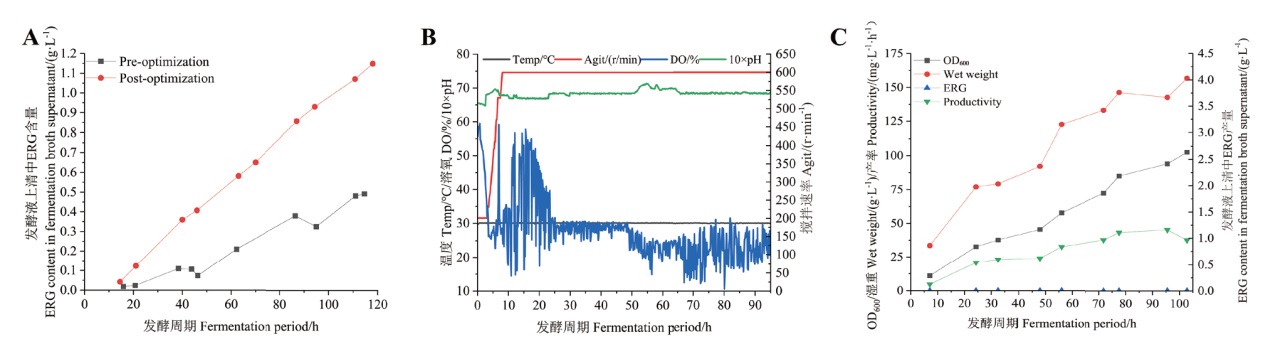
Fig. 6 Fermentation and yield curves A: Comparison of yield before and after optimization of 10 L-bioreactor process. B: Fermentation curves for enlarged fermentation in 30 L-bioreactor. C: Yield curve for fermentation in 30 L-bioreactor
| [1] | Melville DB. Ergothioneine[M]// Vitamins & Hormones. Amsterdam: Elsevier, 1959: 155-204. |
| [2] |
Hunter G. A new test for ergothioneine upon which is based a method for its estimation in simple solution and in blood-filtrates[J]. Biochem J, 1928, 22(1): 4-10.
pmid: 16744012 |
| [3] |
Chen ZH, He YZ, Wu XY, et al. Toward more efficient ergothioneine production using the fungal ergothioneine biosynthetic pathway[J]. Microb Cell Fact, 2022, 21(1): 76.
doi: 10.1186/s12934-022-01807-3 pmid: 35525939 |
| [4] |
Chen L, Zhang LP, Ye XJ, et al. Ergothioneine and its congeners: anti-ageing mechanisms and pharmacophore biosynthesis[J]. Protein Cell, 2024, 15(3): 191-206.
doi: 10.1093/procel/pwad048 |
| [5] | Duan R, Pan HT, Li DC, et al. Ergothioneine improves myocardial remodeling and heart function after acute myocardial infarction via S-glutathionylation through the NF-ĸB dependent Wnt5a-sFlt-1 pathway[J]. Eur J Pharmacol, 2023, 950: 175759. |
| [6] | Mayayo-Vallverdú C, López de Heredia M, Prat E, et al. The antioxidant L-Ergothioneine prevents cystine lithiasis in the Slc7a9-/- mouse model of cystinuria[J]. Redox Biol, 2023, 64: 102801. |
| [7] | Wang Z, Ma JW, Miao ZM, et al. Ergothioneine inhibits the progression of osteoarthritis via the Sirt6/NF-κB axis both in vitro and in vivo[J]. Int Immunopharmacol, 2023, 119: 110211. |
| [8] | Chen Q, Zhou RR, Yang C, et al. Ergothioneine attenuates varicocele-induced testicular damage by upregulating HSP90AA1 in rats[J]. J Biochem Mol Toxicol, 2023, 37(4): e23301. |
| [9] | Leow DMK, Cheah IKM, Fong ZWJ, et al. Protective effect of ergothioneine against 7-ketocholesterol-induced mitochondrial damage in hCMEC/D3 human brain endothelial cells[J]. Int J Mol Sci, 2023, 24(6): 5498. |
| [10] | Hartmann L, Seebeck FP, Schmalz HG, et al. Isotope-labeled ergothioneine clarifies the mechanism of reaction with singlet oxygen[J]. Free Radic Biol Med, 2023, 198: 12-26. |
| [11] | Iqbal S, Jabeen F, Aslam N, et al. Anti-EMT properties of ergothioneine attenuate lipopolysaccharide-induced oxidative stress-mediated acute lung injury via modulating TGF-β/smad/snail signaling pathway[J]. Hum Exp Toxicol, 2023, 42: 9603271231178015. |
| [12] | Bernardo VS, Torres FF, de Paula CP, et al. Potential cytoprotective and regulatory effects of ergothioneine on gene expression of proteins involved in erythroid adaptation mechanisms and redox pathways in K562 cells[J]. Genes, 2022, 13(12): 2368. |
| [13] |
Nakamichi N, Tsuzuku S, Shibagaki F. Ergothioneine and central nervous system diseases[J]. Neurochem Res, 2022, 47(9): 2513-2521.
doi: 10.1007/s11064-022-03665-2 pmid: 35788879 |
| [14] |
Alamgir KM, Masuda S, Fujitani Y, et al. Production of ergothioneine by Methylobacterium species[J]. Front Microbiol, 2015, 6: 1185.
doi: 10.3389/fmicb.2015.01185 pmid: 26579093 |
| [15] | Pluskal T, Ueno M, Yanagida M. Genetic and metabolomic dissection of the ergothioneine and selenoneine biosynthetic pathway in the fission yeast, S. pombe, and construction of an overproduction system[J]. PLoS One, 2014, 9(5): e97774. |
| [16] |
Genghof DS, Van Damme O. Biosynthesis of ergothioneine from endogenous hercynine in Mycobacterium smegmatis[J]. J Bacteriol, 1968, 95(2): 340-344.
doi: 10.1128/jb.95.2.340-344.1968 pmid: 5644441 |
| [17] |
Genghof DS, Inamine E, Kovalenko V, et al. Ergothioneine in microorganisms[J]. J Biol Chem, 1956, 223(1): 9-17.
pmid: 13376573 |
| [18] | 李亚欢. 杏鲍菇中麦角硫因的提取纯化和抗氧化活性研究[D]. 广州: 华南农业大学, 2016. |
| Li YH. Study on extraction, purification and antioxidant activity of ergothionine from Pleurotus eryngii and its antioxidant activity[D]. Guangzhou: South China Agricultural University, 2016. | |
| [19] | Xu J, Yadan JC. ChemInform Abstract: synthesis of L-(+)-ergothioneine[J]. ChemInform, 1996, 27(9): no. |
| [20] | Seebeck FP. In vitro reconstitution of mycobacterial ergothioneine biosynthesis[J]. J Am Chem Soc, 2010, 132(19): 6632-6633. |
| [21] | Qiu YB, Chen ZL, Su EZ, et al. Recent strategies for the biosynthesis of ergothioneine[J]. J Agric Food Chem, 2021, 69(46): 13682-13690. |
| [22] | 刘琦, 毛雨丰, 廖小平, 等. 麦角硫因生物合成研究的新进展[J]. 生物工程学报, 2022, 38(4): 1408-1420. |
| Liu Q, Mao YF, Liao XP, et al. Recent progress in ergothioneine biosynthesis: a review[J]. Chin J Biotechnol, 2022, 38(4): 1408-1420. | |
| [23] | Tanaka N, Kawano Y, Satoh Y, et al. Gram-scale fermentative production of ergothioneine driven by overproduction of cysteine in Escherichia coli[J]. Sci Rep, 2019, 9(1): 1895. |
| [24] | Kamide T, Takusagawa S, Tanaka N, et al. High Production of ergothioneine in Escherichia coli using the sulfoxide synthase from Methylobacterium strains[J]. J Agric Food Chem, 2020, 68(23): 6390-6394. |
| [25] | Zhang HF, Zhang YF, Zhao M, et al. Fermentative production of ergothioneine by exploring novel biosynthetic pathway and remodulating precursor synthesis pathways[J]. J Agric Food Chem, 2024, 72(25): 14264-14273. |
| [26] | Hirasawa T, Shimoyamada Y, Tachikawa Y, et al. Ergothioneine production by Corynebacterium glutamicum harboring heterologous biosynthesis pathways[J]. J Biosci Bioeng, 2023, 135(1): 25-33. |
| [27] | Liu MS, Wu JJ, Yue MY, et al. YaliCMulti and YaliHMulti: stable, efficient multi-copy integration tools for engineering Yarrowia lipolytica[J]. Metab Eng, 2024, 82: 29-40. |
| [28] | van der Hoek SA, Rusnák M, Jacobsen IH, et al. Engineering ergothioneine production in Yarrowia lipolytica[J]. FEBS Lett, 2022, 596(10): 1356-1364. |
| [29] | 陈佳敏. 大肠杆菌麦角硫因合成体系的构建与优化[D]. 无锡: 江南大学, 2023. |
| Chen JM. Construction and optimization of ergothionine synthesis system for Escherichia coli[D]. Wuxi: Jiangnan University, 2023. | |
| [30] | Zhang LW, Tang JW, Feng MQ, et al. Engineering methyltransferase and sulfoxide synthase for high-yield production of ergothioneine[J]. J Agric Food Chem, 2023, 71(1): 671-679. |
| [31] | 王丽, 王阳, 李江华, 等. 产麦角硫因大肠杆菌工程菌株的构建与优化[J]. 生物工程学报, 2022, 38(2): 796-806. |
| Wang L, Wang Y, Li JH, et al. Construction and optimization of ergothioneine-producing Escherichia coli[J]. Chin J Biotechnol, 2022, 38(2): 796-806. | |
| [32] | 陈佳敏, 王阳, 堵国成, 等. 优化前体供给与细胞膜通透性强化大肠杆菌合成麦角硫因[J]. 食品与生物技术学报, 2022, 41(8): 43-52. |
| Chen JM, Wang Y, Du GC, et al. Enhancement of ergothioneine synthesis in Escherichia coli via optimization of precursor supply and cell membrane permeability[J]. J Food Sci Biotechnol, 2022, 41(8): 43-52. | |
| [33] |
Jones GW, Doyle S, Fitzpatrick DA. The evolutionary history of the genes involved in the biosynthesis of the antioxidant ergothioneine[J]. Gene, 2014, 549(1): 161-170.
doi: 10.1016/j.gene.2014.07.065 pmid: 25068406 |
| [34] | Genghof DS, Vandamme O. Biosynthesis of ergothioneine and hercynine by mycobacteria[J]. J Bacteriol, 1964, 87(4): 852-862. |
| [35] | Xiong LB, Xie ZY, Ke J, et al. Engineering Mycolicibacterium neoaurum for the production of antioxidant ergothioneine[J]. Food Bioeng, 2022, 1(1): 26-36. |
| [36] | 刘琦, 张维亚, 姜文侠. 麦角硫因的合成与降解代谢[J]. 天然产物研究与开发, 2015, 27(6): 1112-1117, 1002. |
|
Liu Q, Zhang WY, Jiang WX. Biosynthesis and catabolism of L-ergothioneine[J]. Nat Prod Res Dev, 2015, 27(6): 1112-1117, 1002.
doi: 10.16333/j.1001-6880.2015.06.031 |
|
| [37] | Studier FW. Protein production by auto-induction in high density shaking cultures[J]. Protein Expr Purif, 2005, 41(1): 207-234. |
| [38] | Tahara N, Tachibana I, Takeo K, et al. Boosting auto-induction of recombinant proteins in Escherichia coli with glucose and lactose additives[J]. Protein Pept Lett, 2021, 28(10): 1180-1190. |
| [39] | Crowley EL, Rafferty SP. Review of lactose-driven auto-induction expression of isotope-labelled proteins[J]. Protein Expr Purif, 2019, 157: 70-85. |
| [40] | Fathi-Roudsari M, Maghsoudi N, Maghsoudi A, et al. Auto-induction for high level production of biologically active reteplase in Escherichia coli[J]. Protein Expr Purif, 2018, 151: 18-22. |
| [41] |
EL-Baky NA, Linjawi MH, Redwan EM. Auto-induction expression of human consensus interferon-alpha in Escherichia coli[J]. BMC Biotechnol, 2015, 15: 14.
doi: 10.1186/s12896-015-0128-x pmid: 25886839 |
| [42] | Studier FW. Stable expression clones and auto-induction for protein production in E. coli[J]. Methods Mol Biol, 2014, 1091: 17-32. |
| [43] | Yu YH, Pan HY, Guo LQ, et al. Successful biosynthesis of natural antioxidant ergothioneine in Saccharomyces cerevisiae required only two genes from Grifola frondosa[J]. Microb Cell Fact, 2020, 19(1): 164. |
| [44] |
Kulis-Horn RK, Persicke M, Kalinowski J. Corynebacterium glutamicum ATP-phosphoribosyl transferases suitable for L-histidine production—Strategies for the elimination of feedback inhibition[J]. J Biotechnol, 2015, 206: 26-37.
doi: 10.1016/j.jbiotec.2015.04.001 pmid: 25892668 |
| [45] |
Nakatani T, Ohtsu I, Nonaka G, et al. Enhancement of thioredoxin/glutaredoxin-mediated L-cysteine synthesis from S-sulfocysteine increases L-cysteine production in Escherichia coli[J]. Microb Cell Fact, 2012, 11: 62.
doi: 10.1186/1475-2859-11-62 pmid: 22607201 |
| [46] |
Ohtsu I, Wiriyathanawudhiwong N, Morigasaki S, et al. The L-cysteine/L-cystine shuttle system provides reducing equivalents to the periplasm in Escherichia coli[J]. J Biol Chem, 2010, 285(23): 17479-17487.
doi: 10.1074/jbc.M109.081356 pmid: 20351115 |
| [47] |
Konstantinov K, Kishimoto M, Seki T, et al. A balanced DO-stat and its application to the control of acetic acid excretion by recombinant Escherichia coli[J]. Biotechnol Bioeng, 1990, 36(7): 750-758.
pmid: 18597268 |
| [1] | ZHANG Jing-an, HU Xiao-long, CAO Bei-bei, LIAO Min, SHU Chang-long, ZHANG Jie, WANG Kui, CAO Hai-qun. Construction and Characterization of Rapid Visual Expression Vector for Bacillus thuringiensis [J]. Biotechnology Bulletin, 2025, 41(1): 95-102. |
| [2] | SHEN Zhen-hui, CAO Yao, YANG Lin-lei, LUO Xiang-ying, ZI Ling-shan, LU Qing-qing, LI Rong-chun. Cloning and Bioinformatics Analysis of the Ergothioneine Biosynthesis Genes in Naematelia aurantialba and Stereum hirsutum [J]. Biotechnology Bulletin, 2024, 40(7): 259-272. |
| [3] | ZHANG Mei-yu, ZHAO Yu-bin, WANG Ling-yun, SONG Yuan-da, ZHAO Xin-he, REN Xiao-jie. Research Progress in the Production of Functional Fatty Acid DHA by Microalga Thraustochytrids [J]. Biotechnology Bulletin, 2024, 40(6): 81-94. |
| [4] | WANG Zhou, YU Jie, WANG Jin-hua, WANG Yong-ze, ZHAO Xiao. Anaerobic Expression of Lactate Dehydrogenase to Improve the D-lactic Acid Optical Purity Procluced by Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(5): 290-299. |
| [5] | ZHUANG Ke, LIANG Zhi-xuan, HE Ying-ting, XIE Qiu-ling. Transfer of Antibiotic-resistance Gene AmpR by Escherichia coli DH5α Through Outer Membrane Vesicles [J]. Biotechnology Bulletin, 2024, 40(12): 275-281. |
| [6] | HE Si-cheng, ZHANG Zi-yuan, HAN Yu-qing, MIAO Lin, ZHANG Cui-ying, YU Ai-qun. Research Progress in the Production of Polyunsaturated Fatty Acids by Yarrowia lipolytica Cell Factories [J]. Biotechnology Bulletin, 2024, 40(1): 72-85. |
| [7] | LI Liang, XU Shan-shan, JIANG Yan-jun. Research Progress in the Production of Ergothioneine by Biosynthesis [J]. Biotechnology Bulletin, 2024, 40(1): 86-99. |
| [8] | YANG Hong-yan, HAN Xiao, YANG Jian-jun. Scaling up Production of pDNA Plasmids in Disposable Bioreactors [J]. Biotechnology Bulletin, 2024, 40(1): 168-175. |
| [9] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [10] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [11] | ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering [J]. Biotechnology Bulletin, 2023, 39(8): 137-147. |
| [12] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [13] | CHEN Cai-ping, REN Hao, LONG Teng-fei, HE Bing, LU Zhao-xiang, SUN Jian. Research Advances in the Treatment of Inflammation Bowel Disease Using Escherichia coli Nissle 1917 [J]. Biotechnology Bulletin, 2023, 39(6): 109-118. |
| [14] | YU Hui-li, LI Ai-tao. Application of Cytochrome P450 in the Biosynthesis of Flavors and Fragrances [J]. Biotechnology Bulletin, 2023, 39(4): 24-37. |
| [15] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||