








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (4): 123-133.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0877
LI Wen-lan1( ), HOU Xin-wei1, LI Yan2, ZHAO Rui-jun2, MENG Zhao-dong1, YUE Run-qing1(
), HOU Xin-wei1, LI Yan2, ZHAO Rui-jun2, MENG Zhao-dong1, YUE Run-qing1( )
)
Received:2024-09-09
Online:2025-04-26
Published:2025-04-25
Contact:
YUE Run-qing
E-mail:liwenlantutu@126.com;yuerunqing@126.com
LI Wen-lan, HOU Xin-wei, LI Yan, ZHAO Rui-jun, MENG Zhao-dong, YUE Run-qing. Identification and Resistance Detection of Homozygous and Heterozygous Plants of Transgenic Maize LD05 with Resistances to Insect and Herbicide[J]. Biotechnology Bulletin, 2025, 41(4): 123-133.
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 引物名称 Primer name | 引物序列 Primer sequence (5'-3') |
|---|---|---|---|
| LC913 | AAGTGTTCAGTAAATTATAT | LC953 | TGGCGTTACCCAACTTAATC |
| LC915 | ATACGCCTGTCAAGTGTCAT | LC958 | AATTGGTTCTTGGAATCGCA |
| LC917 | CCTTGTCCTGCATTGCGCAT | LC962 | ATATGTTTCTCAGCGAGCAT |
| LC937 | TCTACTTGGCAAAGGCTTCAGAT | LC964 | CACGGTTCCAGAGGAAAACC |
| LC947 | CAGTAAATTATATACGCCTG | LC966 | GTGCTAGTCAATAGAACTA |
| LC936 | GCACCATCGTCAACCACTACA | LC968 | CACATCGCTAGCTAGTGCTA |
| LC938 | TCTGGCAGCTGGACTTCAGCCTG | LC928 | CGCATAGAAACAACAGAAGTG |
| LC944 | CCAGTACTAAAATCCAGATC | LC929 | TCCTAAAACCAAAATCCAGTA |
| LC948 | CGTCCGCAATGTGTTATTAA | LC933 | GGCAGAGGCATCTTCAACG |
| LC931 | CTTGATGAGACCTGCTGCGT | LC934 | AAACCCCAAGTCCAAGTAAC |
| LC941 | TGAGACCTGCTGCGTAAGCC | zSSIIb-F | CTCCCAATCCTTTGACATCTGC |
| LC951 | AGCTTGGCACTGGCCGTCG | zSSIIb-R | TCGATTTCTCTCTTGGTGACAGG |
Table 1 Sequence information for designing primers
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 引物名称 Primer name | 引物序列 Primer sequence (5'-3') |
|---|---|---|---|
| LC913 | AAGTGTTCAGTAAATTATAT | LC953 | TGGCGTTACCCAACTTAATC |
| LC915 | ATACGCCTGTCAAGTGTCAT | LC958 | AATTGGTTCTTGGAATCGCA |
| LC917 | CCTTGTCCTGCATTGCGCAT | LC962 | ATATGTTTCTCAGCGAGCAT |
| LC937 | TCTACTTGGCAAAGGCTTCAGAT | LC964 | CACGGTTCCAGAGGAAAACC |
| LC947 | CAGTAAATTATATACGCCTG | LC966 | GTGCTAGTCAATAGAACTA |
| LC936 | GCACCATCGTCAACCACTACA | LC968 | CACATCGCTAGCTAGTGCTA |
| LC938 | TCTGGCAGCTGGACTTCAGCCTG | LC928 | CGCATAGAAACAACAGAAGTG |
| LC944 | CCAGTACTAAAATCCAGATC | LC929 | TCCTAAAACCAAAATCCAGTA |
| LC948 | CGTCCGCAATGTGTTATTAA | LC933 | GGCAGAGGCATCTTCAACG |
| LC931 | CTTGATGAGACCTGCTGCGT | LC934 | AAACCCCAAGTCCAAGTAAC |
| LC941 | TGAGACCTGCTGCGTAAGCC | zSSIIb-F | CTCCCAATCCTTTGACATCTGC |
| LC951 | AGCTTGGCACTGGCCGTCG | zSSIIb-R | TCGATTTCTCTCTTGGTGACAGG |
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 引物名称 Primer name | 引物序列 Primer sequence (5'-3') |
|---|---|---|---|
| m2cryAb-vip3A-F | GTTTCCTTTACCGGGGACGA | 18S-F | AAACGGCTACCACATCCAAG |
| m2cryAb-vip3A-A | ACCACCCCCTTCAACTTCAG | 18S-A | CCTCCAATGGATCCTCGTTA |
Table 2 Sequence information of the designed RT-qPCR primers
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 引物名称 Primer name | 引物序列 Primer sequence (5'-3') |
|---|---|---|---|
| m2cryAb-vip3A-F | GTTTCCTTTACCGGGGACGA | 18S-F | AAACGGCTACCACATCCAAG |
| m2cryAb-vip3A-A | ACCACCCCCTTCAACTTCAG | 18S-A | CCTCCAATGGATCCTCGTTA |
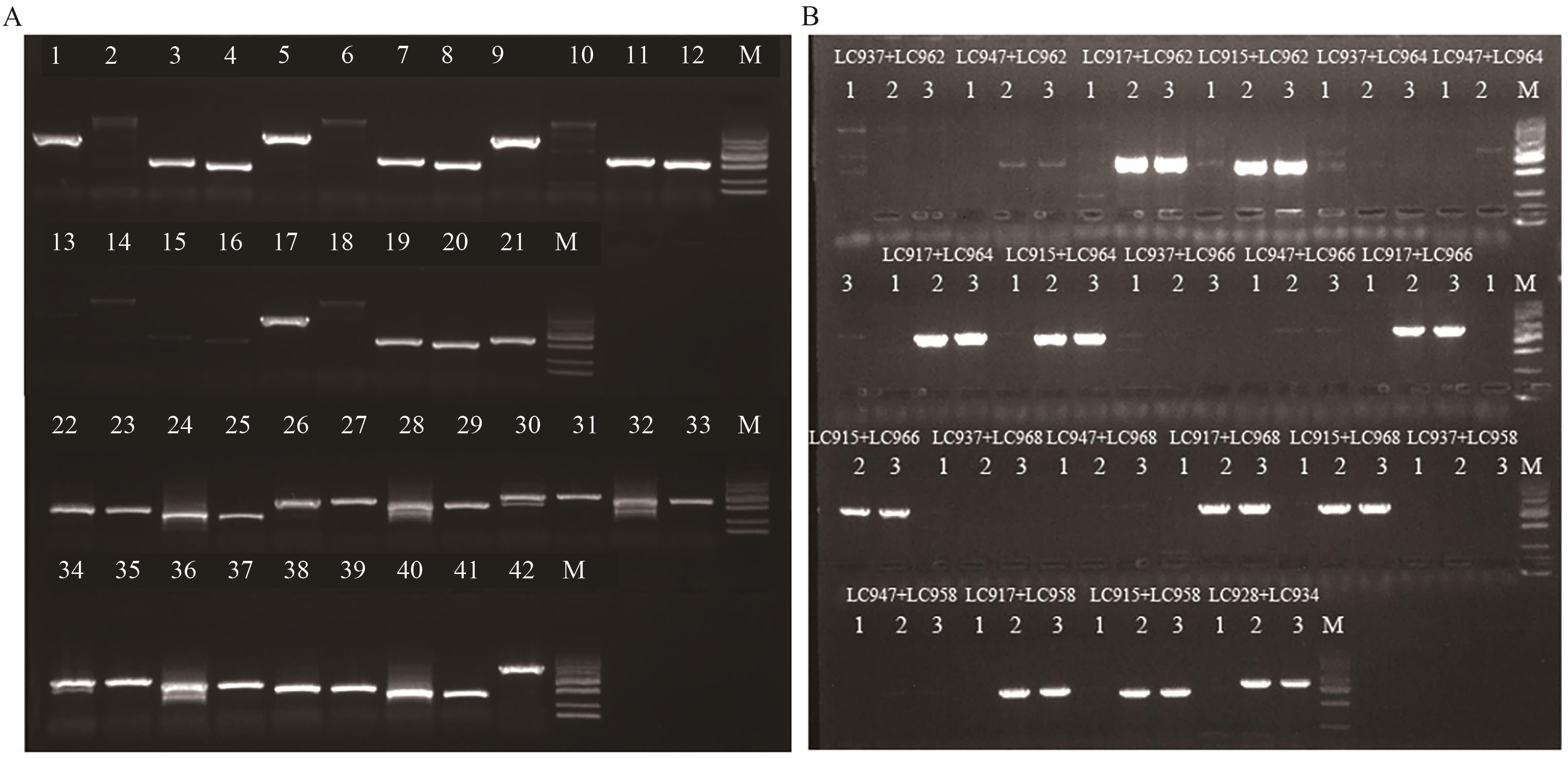
Fig. 2 Screening of primers for homozygous and heterozygous identification of target gene in LD05A: Screening of specific PCR primers for LD05 transformants. M: DL2000 maker. 1: Amplified products of primers LC937 and LC936. 2: Amplified products of primers LC937 and LC938. 3: Amplified products of primers LC937 and LC944. 4: Amplified products of primers LC937 and LC948. 5: Amplified products of primers LC947 and LC936. 6: Amplified products of primers LC947 and LC938. 7: Amplified products of primers LC947 and LC944. 8: Amplified products of primers LC947 and LC948. 9: Amplified products of primers LC917 and LC936. 10: Amplified products of primers LC917 and LC938. 11: Amplified products of primers LC917 and LC944. 12: Amplified products of primers LC917 and LC948. 13: Amplified products of primers LC913 and LC936. 14: Amplified products of primers LC913 and LC938. 15: Amplified products of primers LC913 and LC944. 16: Amplified products of primers LC913 and LC948. 17: Amplified products of primers LC915 and LC936. 18: Amplified products of primers LC915 and LC938. 19: Amplified products of primers LC915 and LC944. 20: Amplified products of primers LC915 and LC948. 21: LC928 and LC929. 22: Amplified products of primers LC931 and LC962. 23: Amplified products of primers LC941 and LC962. 24: Amplified products of primers LC951 and LC962. 25: Amplified products of primers LC953 and LC962. 26: Amplified products of primers LC931 and LC964. 27: Amplified products of primers LC941 and LC964. 28: Amplified products of primers LC951 and LC964. 29: Amplified products of primers LC953 and LC964. 30: Amplified products of primers LC931 and LC966. 31: Amplified products of primers LC941 and LC966. 32: Amplified products of primers LC951 and LC966. 33: Amplified products of primers LC953 and LC966. 34: Amplified products of primers LC931 and LC968. 35: Amplified products of primers LC941 and LC968. 36: Amplified products of primers LC951 and LC968. 37: Amplified products of primers LC953 and LC968. 38: Amplified products of primers LC931 and LC958. 38: Amplified products of primers LC941 and LC958. 40: Amplified products of primers LC951 and LC958. 41: Amplified products of primers LC953 and LC958. 42: Amplified products of primers LC933 and LC934. B: Screening of primers for homozygous and heterozygous identification of target gene in LD05; 1: homozygous DNA sample; 2: heterozygous DNA sample; 3: wild-type DNA sample; M: DL2000 marker, the bands from top to bottom represent 2 000, 1 000, 750, 500, 250, and 100 bp

Fig. 3 Effects of different amplification cycles on amplification efficiency and detection limit test of different primer combinationsA: Effect of different amplification cycles on amplification efficiency; 1, 7, 13, 19, 25, 31: 20 cycles; 2, 8, 14, 20, 26, 32: 22 cycles; 3, 9, 15, 21, 27, 33: 24 cycles; 4, 10, 16, 22, 28, 34: 26 cycles; 5, 11, 17, 23, 29, 35: 28 cycles; 6, 12, 18, 24, 30, 36: 30 cycles; M: DL2000 marker, the bands from top to bottom represent 2 000, 1 000, 750, 500, 250, 100 bp. B: Detection limit test of different primer combinations; 1, 7, 13, 19, 25, 31: template concentration is 100 ng/µL; 2, 8, 14, 20, 26, 32: template concentration of 10 ng/µL; 3, 9, 15, 21, 27, 33: the template concentration is 1 ng/µL; 4, 10, 16, 22, 28, 34: the template concentration is 0.1 ng/µL; 5, 11, 17, 23, 29, 35: the template concentration is 0.01 ng/µL; 6, 12, 18, 24, 30, 36: template concentration is 0.001 ng/µL; M: DL2000 marker

Fig. 4 Analysis of homozygous and heterozygous primersA: PCR amplified results of primer LC915+LC966; B: PCR amplified results of primer LC915+LC948. 1-93: 93 random DNA samples from BC6F2 plants; 94: blank control; 95: negative control; 96: positive control; M: DL2000 marker, the bands from top to bottom represent 2 000, 1 000, 750, 500, 250, and 100 bp

Fig. 5 RT-qPCR and ELISA analysis of m2cryAb-vip3A in homozygous and heterozygous plantsA: RT-qPCR analysis of m2cryAb-vip3A in homozygous and heterozygous plants; B: ELISA analysis of m2cryAb-vip3A encoded protein in homozygous and heterozygous plants. 1, 5, 8, 20: heterozygous plants; 3, 7, 17: homozygous plants. The letters (or alphanumeric combinations) in bars indicate the significant difference in expression analysis by Duncan multiple detection(P<0.05)
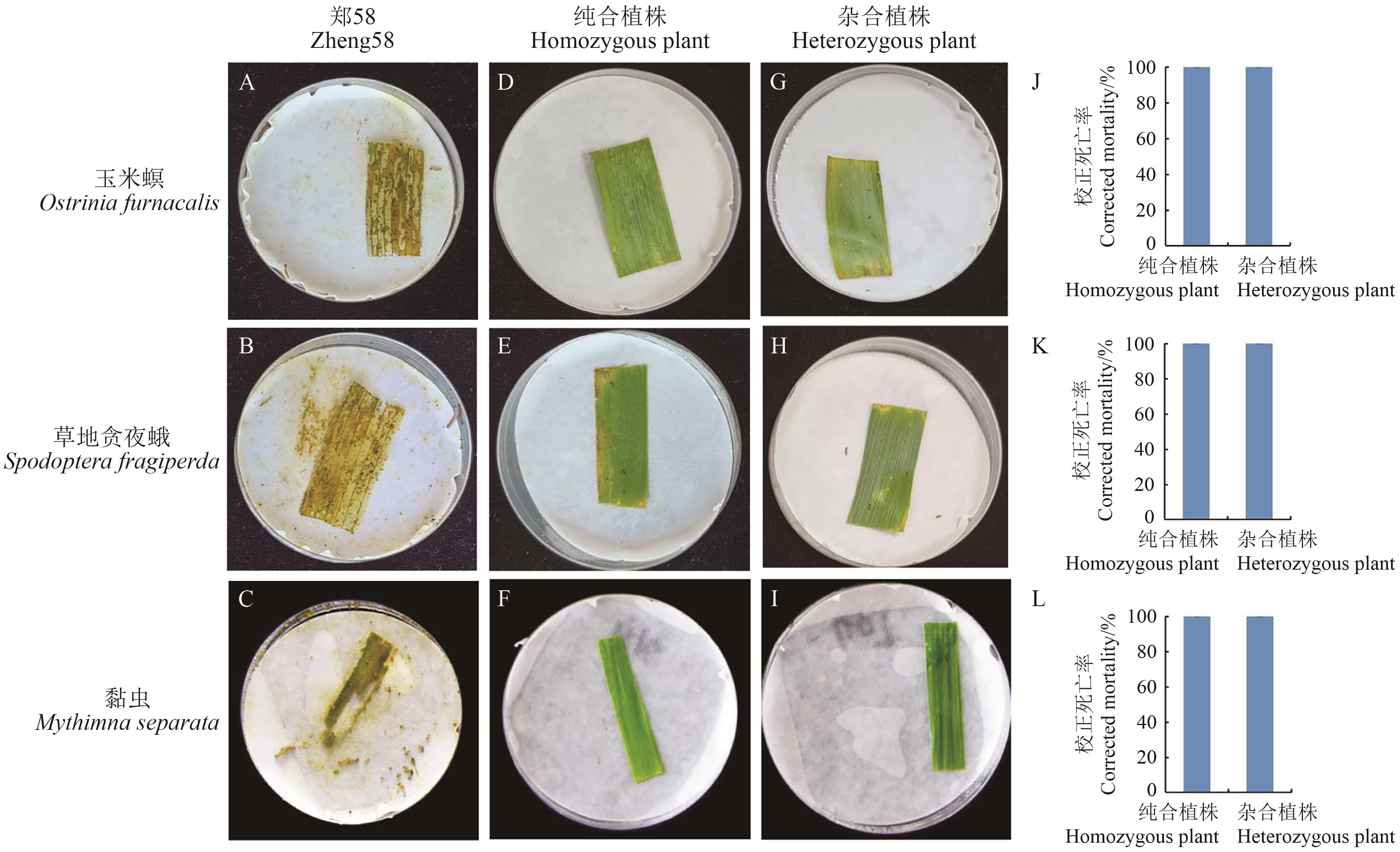
Fig. 6 Results of laboratory bioassay on Ostrinia furnacalis, Spodoptera fragiperda and Mythimna separata at the heart leaf stage (6 d after inoculation)A-C: Zheng 58 leaves at V5 stage; D-F: leaves of homozygous plants at V5 stage; G-I: leaves of heterozygous plants atV5 stage.A, D, G: Ostrinia furnacalis; B, E, H: Spodoptera fragiperda; C, F, I: Mythimna separata; J: corrected mortality of Ostrinia furnacalis; K: corrected mortality of Spodoptera fragiperda; L: corrected mortality of Mythimna separata
靶标害虫 Target pests | 接种时期 Inoculation period | 纯合植株 Homozygous plant | 杂合植株 Heterozygous plant | 郑58 Zheng 58 | ||||
|---|---|---|---|---|---|---|---|---|
级值 Level value | 抗性 Resistance | 级值 Level value | 抗性 Resistance | 级值 Level value | 抗性 Resistance | |||
玉米螟 Ostrinia furnacalis | 心叶期 V5 stage | 1.0 | 高抗 High resistance | 1.0 | 高抗 High resistance | 8.90 | 高感 High sense | |
花丝期 Silking stage | 1.0 | 高抗 High resistance | 1.0 | 高抗 High resistance | 8.99 | 高感 High sense | ||
黏虫 Mythimna separata | 心叶期 V5 stage | 1.01 | 高抗 High resistance | 1.02 | 高抗 High resistance | 8.65 | 高感 High sense | |
棉铃虫 Helicoverpa armigera | 花丝期 Silking stage | 1.0 | 高抗 High resistance | 1.0 | 高抗 High resistance | 6.85 | 感 Sense | |
Table 3 Analysis field trials of Ostrinia furnacalis, Helicoverpa armigera and Helicoverpa armigera
靶标害虫 Target pests | 接种时期 Inoculation period | 纯合植株 Homozygous plant | 杂合植株 Heterozygous plant | 郑58 Zheng 58 | ||||
|---|---|---|---|---|---|---|---|---|
级值 Level value | 抗性 Resistance | 级值 Level value | 抗性 Resistance | 级值 Level value | 抗性 Resistance | |||
玉米螟 Ostrinia furnacalis | 心叶期 V5 stage | 1.0 | 高抗 High resistance | 1.0 | 高抗 High resistance | 8.90 | 高感 High sense | |
花丝期 Silking stage | 1.0 | 高抗 High resistance | 1.0 | 高抗 High resistance | 8.99 | 高感 High sense | ||
黏虫 Mythimna separata | 心叶期 V5 stage | 1.01 | 高抗 High resistance | 1.02 | 高抗 High resistance | 8.65 | 高感 High sense | |
棉铃虫 Helicoverpa armigera | 花丝期 Silking stage | 1.0 | 高抗 High resistance | 1.0 | 高抗 High resistance | 6.85 | 感 Sense | |
| 农艺性状 Agronomic trait | LD05纯合株系 Homozygous plant | 杂合株系 Heterozygous plant | 郑58 Zheng 58 |
|---|---|---|---|
| 抽雄期Tasseling stage/d | 59.7±1.2a | 59.0±1.0a | 59.3±0.6a |
| 生育期Growth stage/d | 109.0±1.0a | 109.3±0.6a | 109.7±1.2a |
| 株高Plant height/cm | 163.7±1.5a | 163.0±4.0a | 164.3±1.5a |
| 穗位高Ear height/cm | 56.3±2.1a | 56.7±0.6a | 56.0±1.0a |
| 穗行数Rows per ear | 12±0a | 12±0a | 12±0a |
| 穗粗Ear diameter/cm | 3.8±0.1a | 3.8±0.0a | 3.8±0.1a |
| 百粒重100-kernel weight/g | 33.2±0.9a | 33.0±0.2a | 33.6±0.7a |
Table 4 Comparative analysis of agronomic characters between LD05 of homozygous and heterozygous plants and Zheng 58
| 农艺性状 Agronomic trait | LD05纯合株系 Homozygous plant | 杂合株系 Heterozygous plant | 郑58 Zheng 58 |
|---|---|---|---|
| 抽雄期Tasseling stage/d | 59.7±1.2a | 59.0±1.0a | 59.3±0.6a |
| 生育期Growth stage/d | 109.0±1.0a | 109.3±0.6a | 109.7±1.2a |
| 株高Plant height/cm | 163.7±1.5a | 163.0±4.0a | 164.3±1.5a |
| 穗位高Ear height/cm | 56.3±2.1a | 56.7±0.6a | 56.0±1.0a |
| 穗行数Rows per ear | 12±0a | 12±0a | 12±0a |
| 穗粗Ear diameter/cm | 3.8±0.1a | 3.8±0.0a | 3.8±0.1a |
| 百粒重100-kernel weight/g | 33.2±0.9a | 33.0±0.2a | 33.6±0.7a |
| 1 | 李浩辉, 刘彩月, 张海文, 等. 2022年度全球转基因作物产业化发展现状及趋势分析 [J]. 中国农业科技导报, 2023, 25(12): 6-16. |
| Li HH, Liu CY, Zhang HW, et al. Global genetically modified crop industrialization trends in 2022 [J]. J Agric Sci Technol, 2023, 25(12): 6-16. | |
| 2 | 李婷婷, 李文娟. 我国玉米空间格局演变及其影响因素研究进展 [J]. 中国农业资源与区划, 2021, 42(2): 87-95. |
| Li TT, Li WJ. Research progress on the evolition of maize spatial pattern and its influencing factors in China [J]. Chin J Agric Resour Reg Plan, 2021, 42(2): 87-95. | |
| 3 | Zhao SY, Yang XM, Liu DZ, et al. Performance of the domestic Bt corn event expressing pyramided Cry1Ab and Vip3Aa19 against the invasive Spodoptera frugiperda (J. E. Smith) in China [J]. Pest Manag Sci, 2023, 79(3): 1018-1029. |
| 4 | 张思雨, 林朝阳, 叶雨轩, 等. 转cry1Ab-vip3Af2和cp4-epsps基因的抗虫耐除草剂水稻的研究 [J]. 浙江农业学报, 2023, 35(8): 1823-1833. |
| Zhang SY, Lin CY, Ye YX, et al. Characterization of transgenic insect resistance and glyphosate tolerance rice expressing cry1Ab-vip3Af2 and cp4-epsps [J]. Acta Agric Zhejiangensis, 2023, 35(8): 1823-1833. | |
| 5 | 林海燕. 表达Cry-VIP3和CP4EPSPS蛋白的转基因抗虫耐草甘膦豇豆的研究 [D]. 杭州: 浙江大学, 2022. |
| Lin HY. A transgenic insect resistance and herbicide tolerance cowpea expression Cry-VIP3 and CP4EPSPS [D]. Hangzhou: Zhejiang University, 2022. | |
| 6 | 黎裕, 王天宇. 玉米转基因技术研发与应用现状及展望 [J]. 玉米科学, 2018, 26(2): 1-15, 22. |
| Li Y, Wang TY. Germplasm enhancement in maize: advances and prospects [J]. J Maize Sci, 2018, 26(2): 1-15, 22. | |
| 7 | 孙伟, 张江丽, 黄丛林, 等. 玉米遗传转化方法的研究进展 [J]. 安徽农业科学, 2007, 35(25): 7772-7774. |
| Sun W, Zhang JL, Huang CL, et al. Progresses in maize genetic transformation techniques [J]. J Anhui Agric Sci, 2007, 35(25): 7772-7774. | |
| 8 | 张元昶, 张首国, 李振科, 等. 转基因玉米遗传转化的研究进展 [J]. 重庆工商大学学报: 自然科学版, 2006, 23(5): 473-476, 495. |
| Zhang YC, Zhang SG, Li ZK, et al. Research progress in genetic transformation methods of transgenic maize [J]. J Chongqing Technol Bus Univ Nat Sci Ed, 2006, 23(5): 473-476, 495. | |
| 9 | Ishida Y, Hiei Y, Komari T. Agrobacterium-mediated transformation of maize [J]. Nat Protoc, 2007, 2(7): 1614-1621. |
| 10 | 周洪昌, 宋伟, 王凤格, 等. 玉米丝黑穗病分子标记辅助选择育种中前景引物与背景引物的筛选 [J]. 分子植物育种, 2011, 9(4): 450-456. |
| Zhou HC, Song W, Wang FG, et al. The selection of foreground and background markers used in maize resistance head smut marker assisted breeding [J]. Mol Plant Breed, 2011, 9(4): 450-456. | |
| 11 | German MA, Kandel- Kfir M, Swarzberg D, et al. A rapid method for the analysis of zygosity in transgenic plants [J]. Plant Science, 2003, 164(2): 183-187. |
| 12 | Sridevi G, Parameswari C, Rajamuni P, et al. Identification of hemizygous and homozygous transgenic rice plants in T1 generation by DNA blot analysis [J]. Plant Biotechnol, 2006, 23(5): 531-534. |
| 13 | 贾芝琪, 张忠华, 崔艳红, 等. 利用外源基因侧翼序列扩增筛选番茄纯合转基因植株 [J]. 农业生物技术学报, 2009, 17(5): 820-824. |
| Jia ZQ, Zhang ZH, Cui YH, et al. Screening homozygous transgenic plants by flanking sequences amplification of T-DNA in tomatoes [J]. J Agric Biotechnol, 2009, 17(5): 820-824. | |
| 14 | 张焕春, 汪小福, 李玥莹, 等. 转Cry1Ab水稻纯合体快速准确的PCR鉴定方法 [J]. 浙江农业学报, 2012, 24(4): 549-554. |
| Zhang HC, Wang XF, Li YY, et al. A rapid and accurate PCR method for homozygous lines screening for genetically modified rice containing Cry1Ab [J]. Acta Agric Zhejiangensis, 2012, 24(4): 549-554. | |
| 15 | 岳润清, 李文兰, 孟昭东. 转基因抗虫耐除草剂玉米自交系LG11的获得及抗性分析 [J]. 作物学报, 2024, 50(1): 89-99. |
| Yue RQ, Li WL, Meng ZD. Acquisition and resistance analysis of transgenic Maize Inbred Line LG11 with insect and herbicide resistance [J]. Acta Agron Sin, 2024, 50(1): 89-99. | |
| 16 | 王振营, 王晓鸣. 我国玉米病虫害发生现状、趋势与防控对策 [J]. 植物保护, 2019, 45(1): 1-11. |
| Wang ZY, Wang XM. Current status and management strategies for corn pests and diseases in China [J]. Plant Prot, 2019, 45(1): 1-11. | |
| 17 | 孙红炜, 李凡, 高瑞, 等. 转cry1Ab/cry2Aj和G10evo-epsps基因玉米中Bt蛋白的时空表达及抗性评价 [J]. 生物安全学报, 2018, 27(1): 63-68. |
| Sun HW, Li F, Gao R, et al. Bt protein spatial-temporal expression and evaluation for resistance of transgenic cry1Ab/cry2Aj and G10evo-epsps maize [J]. J Biosaf, 2018, 27(1): 63-68. | |
| 18 | 王振, 高树仁, 王霞, 等. 玉米不育系回交转育群体背景选择效果的研究 [J]. 玉米科学, 2018, 26(2): 23-29. |
| Wang Z, Gao SR, Wang X, et al. Effect of backcross breeding of maize male sterile line on population background selection [J]. J Maize Sci, 2018, 26(2): 23-29. | |
| 19 | 朱少喜, 金肇阳, 葛建镕, 等. 基于KASP平台的转基因玉米高通量特异性检测方法 [J]. 生物技术通报, 2023, 39(6): 133-140. |
| Zhu SX, Jin ZY, Ge JR, et al. High-throughput specific detection methods for transgenic maize based on the KASP platform [J]. Biotechnol Bull, 2023, 39(6): 133-140. | |
| 20 | 朱少喜, 张云龙, 赵怡锟, 等. 基于60K芯片的玉米回交群体精准背景选择策略 [J]. 玉米科学, 2023, 31(4): 43-51. |
| Zhu SX, Zhang YL, Zhao YK, et al. Accurate background selection strategy of maize backcross population based on 60K chip [J]. J Maize Sci, 2023, 31(4): 43-51. | |
| 21 | 石优. 分子标记辅助选择改良玉米自交系小斑病抗性 [D]. 杨凌: 西北农林科技大学, 2023. |
| Shi Y. Molecular marker-assisted selection for improving resistance of maize inbred lines to leaf spot disease [D]. Yangling: Northwest A & F University, 2023. | |
| 22 | Bilbo TR, Reay-Jones FPF, Reisig DD, et al. Development, survival, and feeding behavior of Helicoverpa zea (Lepidoptera: Noctuidae) relative to Bt protein concentrations in corn ear tissues [J]. PLoS One, 2019, 14(8): e0221343. |
| 23 | 李香菊. 我国耐除草剂转基因作物研发与产业化应用前景 [J]. 植物保护, 2023, 49(5): 316-324. |
| Li XJ. Development of herbicide tolerant crops and their commercialization in China [J]. Plant Prot, 2023, 49(5): 316-324. | |
| 24 | 龙丽坤, 赵宁, 夏蔚, 等. 转基因玉米CM8101特异性定性PCR检测方法 [J]. 中国农学通报, 2021, 37(23): 23-28. |
| Long LK, Zhao N, Xia W, et al. CM8101 transgenic maize: qualitative PCR assay detection of specificity [J]. Chin Agric Sci Bull, 2021, 37(23): 23-28. | |
| 25 | 李葱葱, 谢苹, 董立明, 等. 抗虫耐除草剂玉米GH5112E-117C定性PCR检测方法 [J]. 生物技术通报, 2020, 36(5): 64-67. |
| Li CC, Xie P, Dong LM, et al. Qualitative PCR assay for the detection of GH5112E-117C transgenic maize resistant to insects and herbicides [J]. Biotechnol Bull, 2020, 36(5): 64-67. | |
| 26 | 李凌燕, 肖冰, 张旭冬, 等. 转基因耐除草剂玉米MON87419品系特异性定性PCR检测方法的建立 [J]. 江苏农业科学, 2023, 51(1): 50-57. |
| Li LY, Xiao B, Zhang XD, et al. Establishment of specific qualitative PCR detection method for transgenic herbicide-tolerant maize strain MON87419 [J]. Jiangsu Agric Sci, 2023, 51(1): 50-57. | |
| 27 | 白耀宇, 蒋明星, 程家安. Bt水稻Cry1Ab杀虫蛋白表达的时间动态及其在水稻土中的降解 [J]. 生态学报, 2005, 25(7): 1583-1590. |
| Bai YY, Jiang MX, Cheng JA. Temporal expression patterns of Cry1Ab insecticidal protein in Bt rice plants and its degradation in paddy soils [J]. Acta Ecol Sin, 2005, 25(7): 1583-1590. | |
| 28 | 姜志磊, 刘德璞, 李晓辉, 等. 转基因抗虫玉米Bt毒蛋白的时空表达分析 [J]. 吉林农业科学, 2008, 33(6): 35-37. |
| Jiang ZL, Liu DP, Li XH, et al. Studies on the temporal and spatial expressions of bt toxin protein of bt transgenic maize [J]. J Jilin Agric Sci, 2008, 33(6): 35-37. | |
| 29 | Trtikova M, Wikmark OG, Zemp N, et al. Transgene expression and Bt protein content in transgenic Bt maize (MON810) under optimal and stressful environmental conditions [J]. PLoS One, 2015, 10(4): e0123011. |
| 30 | 王冬梅, 李海强, 丁瑞丰, 等. 新疆北部地区转Bt基因棉外源杀虫蛋白表达时空动态研究 [J]. 棉花学报, 2012, 24(1): 18-26. |
| Wang DM, Li HQ, Ding RF, et al. Spatio-temporal expression of foreign bt insecticidal protein in transgenic bt cotton varieties in northern Xinjiang Province, China [J]. Cotton Sci, 2012, 24(1): 18-26. |
| [1] | LIU Tong-tong, LI Xiao-hui, YANG Jun-long, CHEN Wang, YU Meng, WANG Chao-fan, WANG Feng-ru, KE Shao-ying. Functional Study on ZmSTART1 Regulation of Maize Vascular Bundle Formation [J]. Biotechnology Bulletin, 2025, 41(4): 115-122. |
| [2] | WANG Tao, HU She-wei, ZHANG Yu, DENG Wen-wen, SHANG Chun-yuan, WANG Wan-yi. Research Progress in Starch Biosynthesis and Regulatory Factors in Maize Kernel [J]. Biotechnology Bulletin, 2025, 41(3): 1-13. |
| [3] | TIAN Xu-rui, HUO Xin-yi, GUO Yun-han, XIANG Lin, CHAN Zhu-long, WANG Yan-ping. Expression and Functional Analysis of Gene LoSAUR10 from Lilium Oriental Hybrids [J]. Biotechnology Bulletin, 2025, 41(1): 221-229. |
| [4] | HU Jin-jin, LI Su-zhen, MA Xu-hui, LIU Xiao-qing, XIE Shan-shan, JIANG Hai-yang, CHEN Ru-mei. Regulation of Maize Anthocyanin Biosynthesis Metabolism [J]. Biotechnology Bulletin, 2024, 40(6): 34-44. |
| [5] | SUN Ya-nan, WANG Chun-xue, WANG Xin, DU Bing-hai, LIU Kai, WANG Cheng-qiang. Biocontrol Characteristics of Bacillus atrophaeus CNY01 and Its Salt-resistant and Growth-promoting Effect on Maize Seedling [J]. Biotechnology Bulletin, 2024, 40(5): 248-260. |
| [6] | WANG Jia-wei, LI Chen, LIU Jian-li, ZHOU Shi-jie, YI Jia-min, YANG Jin-yuan, KANG Peng. Effects of Endophytic Fungal Inoculation on the Seedling Growth of Silage Maize [J]. Biotechnology Bulletin, 2024, 40(4): 189-202. |
| [7] | PENG Yu-jia, LI Wen-cui, LIU Yong-bo. Research Progress in the Evolution Mechanisms for Insect Resistance to Insecticides and Bt-transgenic Plants [J]. Biotechnology Bulletin, 2024, 40(4): 40-51. |
| [8] | HU Yi-wa, CHEN Lu. Research Advance and Applications in Maize Wild Relatives Genomes [J]. Biotechnology Bulletin, 2024, 40(3): 14-24. |
| [9] | LI Can, JIANG Xiang-ning, GAI Ying. Cloning of the LkF3H2 Gene in Larix kaempferi and Its Function in Regulating Flavonoid Metabolism [J]. Biotechnology Bulletin, 2024, 40(2): 245-252. |
| [10] | YIN Zi-wei, HONG Yu. Study on the Effect of Rhodococcus rhodochrous NB1 on the Tolerance to Salt and Growth-promoting of Maize and Its Whole Genome [J]. Biotechnology Bulletin, 2024, 40(12): 193-207. |
| [11] | TIAN Jin, ZHANG Yue-qiu, ZHANG Hua, CHEN Zi-yan, TIAN Lu, WANG Hao-qian, GAO Fang-rui, LIANG Jin-gang, CHEN Hong. Specific Qualitative PCR Detection Method for Transgenic Maize Zheda Ruifeng 8 [J]. Biotechnology Bulletin, 2024, 40(12): 45-52. |
| [12] | LIU Yi-jun, YAN Wei, HE Yu-xuan, DONG Li-ming, LONG Li-kun, LI Fei-wu. Development and Application of DNA Standard Molecules of Transgenic Soybean Multi-target Plasmid [J]. Biotechnology Bulletin, 2024, 40(11): 169-183. |
| [13] | CHANG Lu-yin, WANG Zhong-hua, LI Feng-min, GAO Zi-yuan, ZHANG Hui-hong, WANG Yi, LI Fang, HAN Yan-lai, JIANG Ying. Screening Multi-functional Rhizobacteria from Maize Rhizosphere and Their Ehancing Effects on Winter Wheat-Summer Maize Rotation System [J]. Biotechnology Bulletin, 2024, 40(1): 231-242. |
| [14] | WANG Bao-bao, WANG Hai-yang. Molecular Design of Ideal Plant Architecture for High-density Tolerance of Maize Plant [J]. Biotechnology Bulletin, 2023, 39(8): 11-30. |
| [15] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||