








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (8): 146-154.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0104
REN Rui-bin1,2( ), SI Er-jing1,2, WAN Guang-you1,2, WANG Jun-cheng1,2, YAO Li-rong1,2, ZHANG Hong1,2, MA Xiao-le1,2, LI Bao-chun1,3, WANG Hua-jun1,2, MENG Ya-xiong1,2(
), SI Er-jing1,2, WAN Guang-you1,2, WANG Jun-cheng1,2, YAO Li-rong1,2, ZHANG Hong1,2, MA Xiao-le1,2, LI Bao-chun1,3, WANG Hua-jun1,2, MENG Ya-xiong1,2( )
)
Received:2025-01-24
Online:2025-08-26
Published:2025-08-14
Contact:
MENG Ya-xiong
E-mail:1318361099@qq.com;yxmeng1@163.com
REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea[J]. Biotechnology Bulletin, 2025, 41(8): 146-154.
| 基因ID Gene ID | 基因 Gene | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) |
|---|---|---|---|
| P. graminea_GLEAN_10001959 | PgGH17-1 | ACCGTCTACCACACCGTCTA | GGGTAGTTAGGAGGAGCGGA |
| P. graminea_GLEAN_10003434 | PgGH17-2 | CCTACGCCTACACCATCACC | CACGGGTCTTTGGGTTGTTG |
| P. graminea_GLEAN_10004508 | PgGH17-3 | AAATGCTTGTCTGAGGCCGA | TGTGGGATCCACGTATGCAC |
| P. graminea_GLEAN_10006809 | PgGH17-4 | GCAATGAACCAGTACCCCGA | CTGAGTCGGCCATGTTGTCT |
| P. graminea_GLEAN_10007745 | PgGH17-5 | TGGACCGTCTACGTCAATGC | AACAGTTTGGTGGTAGGCGT |
| P. graminea_GLEAN_10008820 | PgGH17-6 | AGGCACCGACTATGAAGCTG | TTGAAGCCGTAGTCGAGCAG |
| Pg-Actin | Pg-Actin | GCGGTTACACCTCTCTACCACC | AGTCTGGATCTCCTGCTCAAAG |
Table 1 Primers of PgGH17 gene family used for RT-qPCR
| 基因ID Gene ID | 基因 Gene | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) |
|---|---|---|---|
| P. graminea_GLEAN_10001959 | PgGH17-1 | ACCGTCTACCACACCGTCTA | GGGTAGTTAGGAGGAGCGGA |
| P. graminea_GLEAN_10003434 | PgGH17-2 | CCTACGCCTACACCATCACC | CACGGGTCTTTGGGTTGTTG |
| P. graminea_GLEAN_10004508 | PgGH17-3 | AAATGCTTGTCTGAGGCCGA | TGTGGGATCCACGTATGCAC |
| P. graminea_GLEAN_10006809 | PgGH17-4 | GCAATGAACCAGTACCCCGA | CTGAGTCGGCCATGTTGTCT |
| P. graminea_GLEAN_10007745 | PgGH17-5 | TGGACCGTCTACGTCAATGC | AACAGTTTGGTGGTAGGCGT |
| P. graminea_GLEAN_10008820 | PgGH17-6 | AGGCACCGACTATGAAGCTG | TTGAAGCCGTAGTCGAGCAG |
| Pg-Actin | Pg-Actin | GCGGTTACACCTCTCTACCACC | AGTCTGGATCTCCTGCTCAAAG |
基因 Gene | 氨基酸数 Number of amino acids (aa) | 分子质量 Molecular weight (Da) | 等电点 Isoelectric point | 亲水性平均值 Grand average of hydropathicity (GRAVY) | 不稳定系数 Instability index | 跨膜结构数 Number of transmembrane structures | 脂肪指数 Aliphatic index | 信号肽长度 Length of signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| PgGH17-1 | 530 | 55 200.02 | 5.09 | -0.165 | 48.99 | 0 | 65.96 | 16 | Extracellular matrix |
| PgGH17-2 | 454 | 48 488.13 | 8.66 | -0.105 | 33.78 | 0 | 75.86 | 17 | Extracellular matrix |
| PgGH17-3 | 443 | 46 301.96 | 5.29 | -0.185 | 41.99 | 0 | 75.12 | 22 | Extracellular matrix |
| PgGH17-4 | 648 | 70 125.37 | 5.67 | -0.507 | 40.27 | 1 | 65.43 | 0 | Plasma membrane |
| PgGH17-5 | 320 | 34 680.98 | 5.18 | -0.164 | 14.52 | 0 | 82.00 | 19 | Extracellular matrix |
| PgGH17-6 | 305 | 32 198.21 | 5.06 | -0.125 | 19.54 | 0 | 68.33 | 19 | Extracellular matrix |
Table 2 Information of PgGH17 gene family members in P. graminea
基因 Gene | 氨基酸数 Number of amino acids (aa) | 分子质量 Molecular weight (Da) | 等电点 Isoelectric point | 亲水性平均值 Grand average of hydropathicity (GRAVY) | 不稳定系数 Instability index | 跨膜结构数 Number of transmembrane structures | 脂肪指数 Aliphatic index | 信号肽长度 Length of signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| PgGH17-1 | 530 | 55 200.02 | 5.09 | -0.165 | 48.99 | 0 | 65.96 | 16 | Extracellular matrix |
| PgGH17-2 | 454 | 48 488.13 | 8.66 | -0.105 | 33.78 | 0 | 75.86 | 17 | Extracellular matrix |
| PgGH17-3 | 443 | 46 301.96 | 5.29 | -0.185 | 41.99 | 0 | 75.12 | 22 | Extracellular matrix |
| PgGH17-4 | 648 | 70 125.37 | 5.67 | -0.507 | 40.27 | 1 | 65.43 | 0 | Plasma membrane |
| PgGH17-5 | 320 | 34 680.98 | 5.18 | -0.164 | 14.52 | 0 | 82.00 | 19 | Extracellular matrix |
| PgGH17-6 | 305 | 32 198.21 | 5.06 | -0.125 | 19.54 | 0 | 68.33 | 19 | Extracellular matrix |
| 保守基序 Conserved motifs | 氨基酸序列 Sequence of amino acids | 氨基酸数目 Number of amino acids |
|---|---|---|
| Motif 1 | DJIVGISVGNEDLYR | 15 |
| Motif 2 | GKPVWVAETGWP | 12 |
| Motif 3 | RJYGTDCNQTQNVIQSAIKHAIK | 23 |
| Motif 4 | NFYQDWFCPRLDYGGDYFWFSAFDEPWK | 28 |
| Motif 5 | DIIGANVHPFF | 11 |
| Motif 6 | GFNYGAKWG | 9 |
| Motif 7 | LLLGJWLD | 8 |
| Motif 8 | QANRGMGFPM | 10 |
| Motif 9 | RNQFRG | 6 |
| Motif 10 | RFWDEVGCW | 9 |
Table 3 Information of amino acids in conserved motifs of PgGH17 genes family in P. graminea
| 保守基序 Conserved motifs | 氨基酸序列 Sequence of amino acids | 氨基酸数目 Number of amino acids |
|---|---|---|
| Motif 1 | DJIVGISVGNEDLYR | 15 |
| Motif 2 | GKPVWVAETGWP | 12 |
| Motif 3 | RJYGTDCNQTQNVIQSAIKHAIK | 23 |
| Motif 4 | NFYQDWFCPRLDYGGDYFWFSAFDEPWK | 28 |
| Motif 5 | DIIGANVHPFF | 11 |
| Motif 6 | GFNYGAKWG | 9 |
| Motif 7 | LLLGJWLD | 8 |
| Motif 8 | QANRGMGFPM | 10 |
| Motif 9 | RNQFRG | 6 |
| Motif 10 | RFWDEVGCW | 9 |
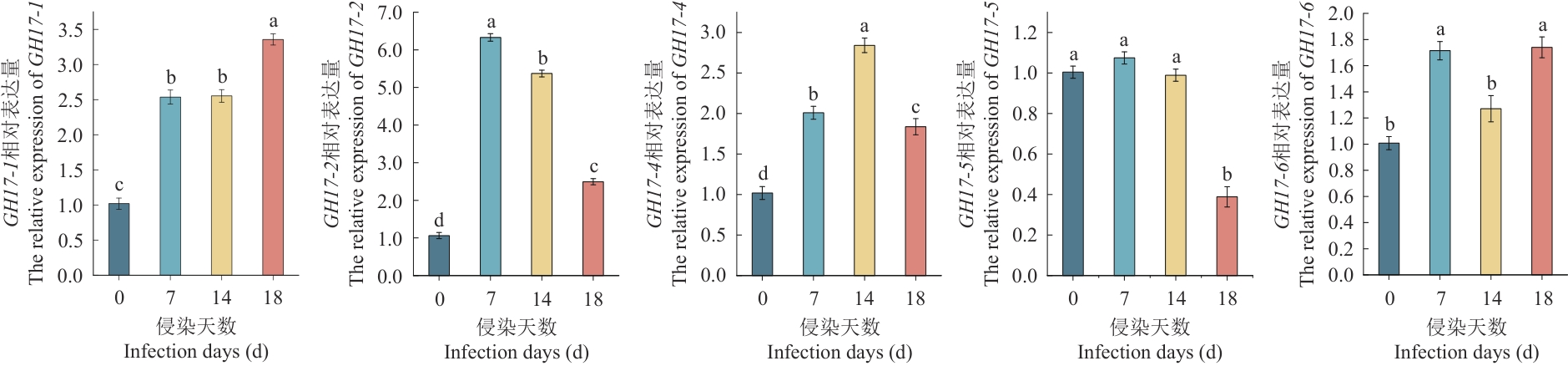
Fig. 4 Variations in the relative expressions of the PgGH17 gene at different infection periodsDifferent lowercase letters indicate significant differences (P<0.05)
| [1] | 侯静静, 何智宏, 司二静, 等. 大麦条纹病原菌的RAPD遗传多样性分析及大麦亲本抗性评价 [J]. 西北农业学报, 2020, 29(4): 512-520. |
| Hou JJ, He ZH, Si EJ, et al. RAPD genetic diversity analysis of Pyrenophora graminea and evaluation of barley parent resistance [J]. Acta Agric Boreali Occidentalis Sin, 2020, 29(4): 512-520. | |
| [2] | Wang Y, Hu Q, Yao YH, et al. Transcriptome, miRNA, and degradome sequencing reveal the leaf stripe (Pyrenophora graminea) resistance genes in Tibetan hulless barley [J]. BMC Plant Biol, 2025, 25(1): 71. |
| [3] | 孟亚雄, 何智宏, 汪军成, 等. 大麦条纹病侵染对大麦叶片蛋白组的影响 [J]. 农业生物技术学报, 2016, 24(6): 824-836. |
| Meng YX, He ZH, Wang JC, et al. Effect on the proteome of barley (Hordeum vulgare) leaves after inoculation of Pyrenophora graminea [J]. J Agric Biotechnol, 2016, 24(6): 824-836. | |
| [4] | 祁天涛, 司二静, 郭铭, 等. 大麦条纹病菌基因Pgr03902致病功能研究 [J]. 植物保护, 2023, 49(6): 16-22, 54. |
| Qi TT, Si EJ, Guo M, et al. Study on the function of the gene Pgr03902 of Pyrenophora graminea [J]. Plant Prot, 2023, 49(6): 16-22, 54. | |
| [5] | 郭铭, 孙莉莎, 司二静, 等. 大麦条纹病病菌对大麦叶片防御酶活性的影响 [J]. 西北农业学报, 2021, 30(2): 295-303. |
| Guo M, Sun LS, Si EJ, et al. Effect of Pyrenophora graminea stress to defensive enzymes activity in barley leaves [J]. Acta Agric Boreali Occidentalis Sin, 2021, 30(2): 295-303. | |
| [6] | Ghannam A, Alek H, Doumani S, et al. Deciphering the transcriptional regulation and spatiotemporal distribution of immunity response in barley to Pyrenophora graminea fungal invasion [J]. BMC Genomics, 2016, 17: 256. |
| [7] | Arabi M, Jawhar M. Heterogeneity in Pyrenophora graminea as revealed by its-rflp [J]. J Plant Pathol, 2007, 89(3): 391-395. |
| [8] | Bakri Y, Arabi ME, Jawhar M. Heterogeneity in the ITS of the ribosomal DNA of Pyrenophora graminea isolates differing in xylanase and amylase production [J]. Mikrobiologiia, 2011, 80(4): 486-489. |
| [9] | Bhuvaneswari V, Srivastava AK, Paul PK. Aqueous fruit extracts of Azadirachta indica induce systemic acquired resistance in barley against Drechslera graminea [J]. Arch Phytopathol Plant Prot, 2012, 45(8): 898-908. |
| [10] | Biselli C, Urso S, Bernardo L, et al. Identification and mapping of the leaf stripe resistance gene Rdg1a in Hordeum spontaneum [J]. Theor Appl Genet, 2010, 120(6): 1207-1218. |
| [11] | Biselli C, Urso S, Tacconi G, et al. Haplotype variability and identification of new functional alleles at the Rdg2a leaf stripe resistance gene locus [J]. Theor Appl Genet, 2013, 126(6): 1575-1586. |
| [12] | 杨瑞, 郑果, 王生荣. 大麦条纹病病原菌的生物学特性研究 [J]. 贵州农业科学, 2010, 38(9): 106-109. |
| Yang R, Zheng G, Wang SR. Biological characteristics of pathogenic fungus causing barley stripe [J]. Guizhou Agric Sci, 2010, 38(9): 106-109. | |
| [13] | 司二静, 张宇, 孟亚雄, 等. 大麦抗条纹病基因的定位分析 [J]. 植物保护学报, 2019, 46(4): 723-729. |
| Si EJ, Zhang Y, Meng YX, et al. Molecular mapping of a gene for resistance to barley leaf stripe [J]. J Plant Prot, 2019, 46(4): 723-729. | |
| [14] | 吴宽然, 杨建明, 朱靖环, 等. 大麦条纹病抗性及防治研究进展 [J]. 浙江农业学报, 2013, 25(4): 903-907. |
| Wu KR, Yang JM, Zhu JH, et al. Advances of research on control of barley leaf stripe disease [J]. Acta Agric Zhejiangensis, 2013, 25(4): 903-907. | |
| [15] | 许佳君, 盛海安, 刘敏. 10%二硫氰基甲烷EC等不同药剂拌种防治大麦条纹病试验 [J]. 上海农业科技, 2012(2): 132. |
| Xu JJ, Sheng HA, Liu M. Experiment on controlling barley stripe disease by dressing seeds with 10% dithiocyanatomethane EC and other different chemicals [J]. Shanghai Agric Sci Technol, 2012(2): 132. | |
| [16] | 陈桂华, 褚金云, 金保根, 等. 金山区大麦条纹病暴发情况及原因分析 [J]. 上海农业科技, 2009(5): 134. |
| Chen GH, Chu JY, Jin BG, et al. Outbreak of barley stripe disease in Jinshan district and its causes analysis [J]. Shanghai Agric Sci Technol, 2009(5): 134. | |
| [17] | 司二静, 杨淑莲, 李葆春, 等. 甘肃省大麦条纹病病原菌致病力分化、rDNA-ITS及遗传多样性分析 [J]. 植物保护学报, 2017, 44(1): 84-92. |
| Si EJ, Yang SL, Li BC, et al. Pathogenic analysis, rDNA-ITS and genetic diversity of Pyrenophora garminea in Gansu Province [J]. J Plant Prot, 2017, 44(1): 84-92. | |
| [18] | 靳生杰, 洪平, 张丽琼. 玉门市大麦条纹病的发生与防治 [J]. 农业科技与信息, 2008(13): 35-36. |
| Jin SJ, Hong P, Zhang LQ. Occurrence and control of barley stripe disease in Yumen city [J]. Agric Sci Technol Inf, 2008(13): 35-36. | |
| [19] | Gow NAR, Lenardon MD. Architecture of the dynamic fungal cell wall [J]. Nat Rev Microbiol, 2023, 21(4): 248-259. |
| [20] | Hopke A, Brown AJP, Hall RA, et al. Dynamic fungal cell wall architecture in stress adaptation and immune evasion [J]. Trends Microbiol, 2018, 26(4): 284-295. |
| [21] | Rafiei V, Vélëz H, Tzelepis G. The role of glycoside hydrolases in phytopathogenic fungi and oomycetes virulence [J]. Int J Mol Sci, 2021, 22(17): 9359. |
| [22] | Minic Z. Physiological roles of plant glycoside hydrolases [J]. Planta, 2008, 227(4): 723-740. |
| [23] | Calderan-Rodrigues MJ, Fonseca JG, Clemente HS, et al. Glycoside hydrolases in plant cell wall proteomes: predicting functions that could be relevant for improving biomass transformation processes [M]//Advances in Biofuels and Bioenergy. London: InTech, 2018. |
| [24] | 房文霞, 金城. 烟曲霉细胞壁的组分、组装及其功能概述 [J]. 菌物学报, 2018, 37(10): 1307-1316. |
| Fang WX, Jin C. The cell wall of Aspergillus fumigatus: composition, biosynthesis and function [J]. Mycosystema, 2018, 37(10): 1307-1316. | |
| [25] | 魏夏森, 余赛男, 张哲一, 等. β-1, 3-葡聚糖酶的结构、功能及应用研究进展 [J]. 食品科学, 2023, 44(15): 269-277. |
| Wei XS, Yu SN, Zhang ZY, et al. Research progress on structure, function and application of β-1, 3-glucanases [J]. Food Sci, 2023, 44(15): 269-277. | |
| [26] | Wan JX, He M, Hou QQ, et al. Cell wall associated immunity in plants [J]. Stress Biol, 2021, 1(1): 3. |
| [27] | 潘凤英, 刘露露, 孙大运, 等. 植物病原菌糖基水解酶基因家族研究进展 [J]. 生物学杂志, 2022, 39(6): 94-100. |
| Pan FY, Liu LL, Sun DY, et al. Research progress on glycoside hydrolases gene family of plant pathogen [J]. J Biol, 2022, 39(6): 94-100. | |
| [28] | Bradley EL, Ökmen B, Doehlemann G, et al. Secreted glycoside hydrolase proteins as effectors and invasion patterns of plant-associated fungi and oomycetes [J]. Front Plant Sci, 2022, 13: 853106. |
| [29] | Aimanianda V, Simenel C, Garnaud C, et al. The dual activity responsible for the elongation and branching of β-(1, 3)-glucan in the fungal cell wall [J]. mBio, 2017, 8(3): e00619-17. |
| [30] | Jiang Y, Chang ZP, Xu Y, et al. Advances in molecular enzymology of β-1, 3-glucanases: a comprehensive review [J]. Int J Biol Macromol, 2024, 279: 135349. |
| [31] | Patel P, Free SJ. Characterization of Neurospora crassa GH16 GH17 and GH72 gene families of cell wall crosslinking enzymes [J]. Cell Surf, 2022, 8: 100073. |
| [32] | Millet N, Latgé JP, Mouyna I. Members of glycosyl-hydrolase family 17 of A. fumigatus differentially affect morphogenesis [J]. J Fungi, 2018, 4(1): 18. |
| [33] | Wei W, Xu LS, Peng H, et al. A fungal extracellular effector inactivates plant polygalacturonase-inhibiting protein [J]. Nat Commun, 2022, 13(1): 2213. |
| [34] | Fang S, Shang XG, He QF, et al. A cell wall-localized β-1, 3-glucanase promotes fiber cell elongation and secondary cell wall deposition [J]. Plant Physiol, 2023, 194(1): 106-123. |
| [35] | Liu H, Lu XL, Li MF, et al. Plant immunity suppression by an exo-β-1, 3-glucanase and an elongation factor 1α of the rice blast fungus [J]. Nat Commun, 2023, 14(1): 5491. |
| [36] | Gu XY, Cao ZY, Li ZQ, et al. Plant immunity suppression by an β-1, 3-glucanase of the maize anthracnose pathogen Colletotrichum graminicola [J]. BMC Plant Biol, 2024, 24(1): 339. |
| [37] | Henrissat B, Davies GJ. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics [J]. Plant Physiol, 2000, 124(4): 1515-1519. |
| [38] | 孔青洋, 张晓龙, 李娜, 等. 单叶蔷薇GRAS转录因子家族鉴定及表达分析 [J]. 生物技术通报, 2025, 41(1): 210-220. |
| Kong QY, Zhang XL, Li N, et al. Identification and expression analysis of GRAS transcription factor family in Rosa persica [J]. Biotechnol Bull, 2025, 41(1): 210-220. | |
| [39] | 李禹欣, 李苗, 杜晓芬, 等. 谷子SiSAP基因家族的鉴定与表达分析 [J]. 生物技术通报, 2025, 41(1): 143-156. |
| Li YX, Li M, Du XF, et al. Identification and expression analysis of SiSAP gene family in foxtail millet (Setaria italica) [J]. Biotechnol Bull, 2025, 41(1): 143-156. | |
| [40] | 郭铭, 张金福, 司二静, 等. 不同来源大麦对条纹病抗性鉴定及遗传多样性分析 [J]. 植物遗传资源学报, 2022, 23(1): 72-82. |
| Guo M, Zhang JF, Si EJ, et al. Resistance identification and genetic diversity analysis of barley genotypes from different sources to barley stripe disease [J]. J Plant Genet Resour, 2022, 23(1): 72-82. | |
| [41] | 戴贤甬. 植物中的免疫系统研究进展 [J]. 农业科学, 2023(4): 326-336. |
| Dai XY. Research progress of immune system in plants [J]. Hans J Agric Sci, 2023(4): 326-336. | |
| [42] | 王小菲, 高文强, 刘建锋, 等. 植物防御策略及其环境驱动机制 [J]. 生态学杂志, 2015, 34(12): 3542-3552. |
| Wang XF, Gao WQ, Liu JF, et al. Plant defensive strategies and environment-driven mechanisms [J]. Chin J Ecol, 2015, 34(12): 3542-3552. | |
| [43] | Kubicek CP, Starr TL, Glass NL. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi [J]. Annu Rev Phytopathol, 2014, 52: 427-451. |
| [44] | Pervaiz T, Liu TH, Fang X, et al. Identification of GH17 gene family in Vitis vinifera and expression analysis of GH17 under various adversities [J]. Physiol Mol Biol Plants, 2021, 27(7): 1423-1436. |
| [45] | Chandrasekar B, Wanke AL, Wawra S, et al. Fungi hijack a ubiquitous plant apoplastic endoglucanase to release a ROS scavenging β-glucan decasaccharide to subvert immune responses [J]. Plant Cell, 2022, 34(7): 2765-2784. |
| [46] | Ökmen B, Bachmann D, de Wit PJGM. A conserved GH17 glycosyl hydrolase from plant pathogenic Dothideomycetes releases a DAMP causing cell death in tomato [J]. Mol Plant Pathol, 2019, 20(12): 1710-1721. |
| [47] | Chen D, Zeng ZH, Yu CY, et al. Genome-wide identification of GH17s family genes and biological function analysis of SlA6 in tomato [J]. Plants, 2024, 13(17): 2443. |
| [48] | Ruel K, Joseleau JP. Involvement of an extracellular glucan sheath during degradation of Populus wood by Phanerochaete chrysosporium [J]. Appl Environ Microbiol, 1991, 57(2): 374-384. |
| [49] | Ezzine A, Chahed H, Hannachi M, et al. Biochemical and molecular characterization of a new glycoside hydrolase family 17 from Sclerotinia sclerotiorum [J]. Journal of New Sciences, 2016, 28(8): 1610-1621. |
| [50] | 刘露露, 曲俊杰, 潘凤英, 等. 葡萄霜霉菌糖基水解酶基因家族的生物信息学分析 [J]. 南方农业学报, 2021, 52(5): 1229-1237. |
| Liu LL, Qu JJ, Pan FY, et al. Bioinformatic analysis of glycoside hydrolases gene family in Plasmopara viticola [J]. J South Agric, 2021, 52(5): 1229-1237. | |
| [51] | 范玉杰, 武晓雄, 董梦迪, 等. 谷子Glycosyl hydrolases family 17鉴定及在逆境胁迫下的表达分析 [J]. 核农学报, 2024, 38(9): 1660-1673. |
| Fan YJ, Wu XX, Dong MD, et al. Identification of glycosyl hydrolases family 17 in foxtail millet and expression analysis under adversity stress [J]. J Nucl Agric Sci, 2024, 38(9): 1660-1673. | |
| [52] | Shi ZW, Wu JM, Mo HR, et al. Identification of an ethylene-responsive and cell wall-secreting β-1, 3-glucanase, VvGLU1, in the early cell regrowth of grape winter buds triggered by exogenous dormancy releasers [J]. BMC Biol, 2025, 23(1): 22. |
| [53] | 侯静静. 大麦条纹病菌致病性候选基因Pgmiox功能研究 [D]. 兰州: 甘肃农业大学, 2020. |
| Hou JJ. Study on the function of Pgmiox, a candidate gene for pathogenicity of barley stripe disease [D]. Lanzhou: Gansu Agricultural University, 2020. |
| [1] | CHENG Xue, FU Ying, CHAI Xiao-jiao, WANG Hong-yan, DENG Xin. Identification of LHC Gene Family in Setaria italica and Expression Analysis under Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(8): 102-114. |
| [2] | LI Xin-ni, LI Jun-yi, MA Xue-hua, HE Wei, LI Jia-li, YU Jia, CAO Xiao-ning, QIAO Zhi-jun, LIU Si-chen. Identification of the PMEI Gene Family of Pectin Methylesterase Inhibitor in Foxtail Millet and Analysis of Its Response to Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(7): 150-163. |
| [3] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [4] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [5] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [6] | ZHANG Ze, YANG Xiu-li, NING Dong-xian. Identification of 4CL Gene Family in Arachis hypogaea L. and Expression Analysis in Response to Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(7): 117-127. |
| [7] | QU Mei-ling, ZHOU Si-min, ZHANG Jing-yu, HE Jia-wei, ZHU Jia-yuan, LIU Xiao-rong, TONG Qiao-zhen, ZHOU Ri-bao, LIU Xiang-dan. Identification and Expression Analysis of bHLH Transcription Gene Family in Lonicera macranthoides [J]. Biotechnology Bulletin, 2025, 41(6): 256-268. |
| [8] | HUANG Dan, PENG Bing-yang, ZHANG Pan-pan, JIAO Yue, LYU Jia-bin. Identification of HD-Zip Gene Family in Camellia oleifera and Analysis of Its Expression under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(6): 191-207. |
| [9] | WANG Yi-min, LI Ying, DONG Hai-tao, ZHANG Heng-rui, CHANG Lu, GAO Tian-tian, HAN De-jun, WU Jian-hui. Evolutionary Patterns of SRO Family Proteins in the Polyploidization Process of Wheat [J]. Biotechnology Bulletin, 2025, 41(5): 70-81. |
| [10] | HE Wei, LI Jun-yi, LI Xin-ni, MA Xue-hua, XING Yuan, CAO Xiao-ning, QIAO Zhi-ju, LIU Si-chen. Genome-wide Identification of U-box E3 Ubiquitin Ligase Gene Family in Setaria italica and Response Analysis to Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(5): 104-118. |
| [11] | LUO Si-fang, ZHANG Zu-ming, XIE Li-fang, GUO Zi-jing, CHEN Zhao-xing, YANG Yue-hua, YAN Xiang, ZHANG Hong-ming. Genome-wide Identification of GATA Gene Family of Jindou Kumquat (Fortunella hindsii) and Their Expression Analysis in Fruit Development [J]. Biotechnology Bulletin, 2025, 41(5): 218-230. |
| [12] | LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 141-152. |
| [13] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [14] | WANG Tian-tian, CHANG Xue-rui, HUANG Wan-yang, HUANG Jia-xin, MIAO Ru-yi, LIANG Yan-ping, WANG Jing. Identification and Analysis of GASA Gene Family in Pepper (Capsicum annuum L.) [J]. Biotechnology Bulletin, 2025, 41(4): 166-175. |
| [15] | SONG Jia-yi, SU Yun-li, ZHENG Xing-yan, XIA Wen-nian, YANG Dong-mei, HU Hui-zhen. Identification of the Snapdragon Expansin Gene Family and Screening of Its Genes Related to Resistance to Sclerotinia sclerotiorum [J]. Biotechnology Bulletin, 2025, 41(4): 227-242. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||