








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (8): 211-219.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1233
HUA Wen-ping1,2( ), LIU Fei1, HAO Jia-xin1, CHEN Chen2,3
), LIU Fei1, HAO Jia-xin1, CHEN Chen2,3
Received:2024-12-20
Online:2025-08-26
Published:2025-08-14
Contact:
HUA Wen-ping
E-mail:huawenping@126.com
HUA Wen-ping, LIU Fei, HAO Jia-xin, CHEN Chen. Identification and Expression Patterns Analysis of ADH Gene Family in Salvia miltiorrhiza[J]. Biotechnology Bulletin, 2025, 41(8): 211-219.
| 基因Gene | 引物序列 Primer sequence (5′-3′) |
|---|---|
| SmActin | F-AGGAACCACCGATCCAGACA |
| (DQ243702) | R-GGTGCCCTGAGGTCCTGTT |
| SmADH2 | F-GAACCCAAAAGACTACAGCAAGCC R-ACGCCAACAAGAACAGCCACTC |
| SmADH5 | F-GTGTGATGCCAGCAAACGGATT R-TGACGGCGACGGAGTTACCA |
| SmADH6 | F-ATGGGAGGTCTTGCTGAGTTTTGT R-CAATGGCATCACCAGGACGC |
Table 1 Primers used for RT-qPCR
| 基因Gene | 引物序列 Primer sequence (5′-3′) |
|---|---|
| SmActin | F-AGGAACCACCGATCCAGACA |
| (DQ243702) | R-GGTGCCCTGAGGTCCTGTT |
| SmADH2 | F-GAACCCAAAAGACTACAGCAAGCC R-ACGCCAACAAGAACAGCCACTC |
| SmADH5 | F-GTGTGATGCCAGCAAACGGATT R-TGACGGCGACGGAGTTACCA |
| SmADH6 | F-ATGGGAGGTCTTGCTGAGTTTTGT R-CAATGGCATCACCAGGACGC |
| 基因Gene | 基长度 Gene length (bp) | 编码序列长度CDs length (bp) | 内含子数目 Number of introns | 氨基酸数目Number of amino acids | 理论等电点pI | 分子量Molecular weight/kD | 平均亲水性指数 GRAVY | 亚细胞定位Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| SmADH1 | 2 392 | 1 149 | 9 | 382 | 6.38 | 41.32 | -0.014 | Cytoplasm |
| SmADH2 | 2 279 | 1 140 | 8 | 379 | 5.98 | 40.87 | 0.101 | Cytoplasm |
| SmADH3 | 8 279 | 1 149 | 8 | 382 | 6.87 | 41.55 | 0.259 | Cytoplasm |
| SmADH4 | 2 824 | 1 167 | 9 | 388 | 5.75 | 41.83 | 0.137 | Cytoplasm |
| SmADH5 | 4 670 | 1 299 | 7 | 432 | 8.67 | 46.20 | -0.059 | Chloroplasts |
| SmADH6 | 4 849 | 1 317 | 7 | 438 | 8.74 | 46.89 | -0.056 | Chloroplasts |
| SmADH7 | 3 827 | 1 140 | 8 | 379 | 6.37 | 40.79 | 0.063 | Cytoplasm |
Table 2 Properties and locations of the predicted SmADH proteins in Salvia miltiorrhiza
| 基因Gene | 基长度 Gene length (bp) | 编码序列长度CDs length (bp) | 内含子数目 Number of introns | 氨基酸数目Number of amino acids | 理论等电点pI | 分子量Molecular weight/kD | 平均亲水性指数 GRAVY | 亚细胞定位Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| SmADH1 | 2 392 | 1 149 | 9 | 382 | 6.38 | 41.32 | -0.014 | Cytoplasm |
| SmADH2 | 2 279 | 1 140 | 8 | 379 | 5.98 | 40.87 | 0.101 | Cytoplasm |
| SmADH3 | 8 279 | 1 149 | 8 | 382 | 6.87 | 41.55 | 0.259 | Cytoplasm |
| SmADH4 | 2 824 | 1 167 | 9 | 388 | 5.75 | 41.83 | 0.137 | Cytoplasm |
| SmADH5 | 4 670 | 1 299 | 7 | 432 | 8.67 | 46.20 | -0.059 | Chloroplasts |
| SmADH6 | 4 849 | 1 317 | 7 | 438 | 8.74 | 46.89 | -0.056 | Chloroplasts |
| SmADH7 | 3 827 | 1 140 | 8 | 379 | 6.37 | 40.79 | 0.063 | Cytoplasm |
| miRNA | 靶基因 Target gene | 期望值Expectation | 靶位点Target site (bp) | 抑制方式 Inhibition way |
|---|---|---|---|---|
| smi-miR167a | SmADH3 | 4 | 60-80 | 剪切 |
| smi-miR167b | SmADH3 | 4 | 60-80 | 剪切 |
| smi-miR167c | SmADH3 | 4 | 60-80 | 剪切 |
| smi-miR167e | SmADH3 | 4 | 60-80 | 剪切 |
| smi-miRN43 | SmADH4 | 4.5 | 1 079-1 099 | 剪切 |
| smi-miR482c | SmADH5 | 5 | 601-622 | 翻译 |
| smi-miR482d | SmADH5 | 5 | 601-622 | 翻译 |
| smi-miRN18 | SmADH6 | 3.5 | 377-398 | 翻译 |
| smi-miR482c | SmADH6 | 5 | 619-640 | 翻译 |
| smi-miR482d | SmADH6 | 5 | 619-640 | 翻译 |
| smi-miRN5a | SmADH7 | 4.5 | 5-25 | 翻译 |
| smi-miRN5b | SmADH7 | 4.5 | 5-25 | 剪切 |
| smi-miR530a | SmADH7 | 5 | 535-555 | 剪切 |
Table 3 Predicting analysis of miRNA in SmADH genes
| miRNA | 靶基因 Target gene | 期望值Expectation | 靶位点Target site (bp) | 抑制方式 Inhibition way |
|---|---|---|---|---|
| smi-miR167a | SmADH3 | 4 | 60-80 | 剪切 |
| smi-miR167b | SmADH3 | 4 | 60-80 | 剪切 |
| smi-miR167c | SmADH3 | 4 | 60-80 | 剪切 |
| smi-miR167e | SmADH3 | 4 | 60-80 | 剪切 |
| smi-miRN43 | SmADH4 | 4.5 | 1 079-1 099 | 剪切 |
| smi-miR482c | SmADH5 | 5 | 601-622 | 翻译 |
| smi-miR482d | SmADH5 | 5 | 601-622 | 翻译 |
| smi-miRN18 | SmADH6 | 3.5 | 377-398 | 翻译 |
| smi-miR482c | SmADH6 | 5 | 619-640 | 翻译 |
| smi-miR482d | SmADH6 | 5 | 619-640 | 翻译 |
| smi-miRN5a | SmADH7 | 4.5 | 5-25 | 翻译 |
| smi-miRN5b | SmADH7 | 4.5 | 5-25 | 剪切 |
| smi-miR530a | SmADH7 | 5 | 535-555 | 剪切 |
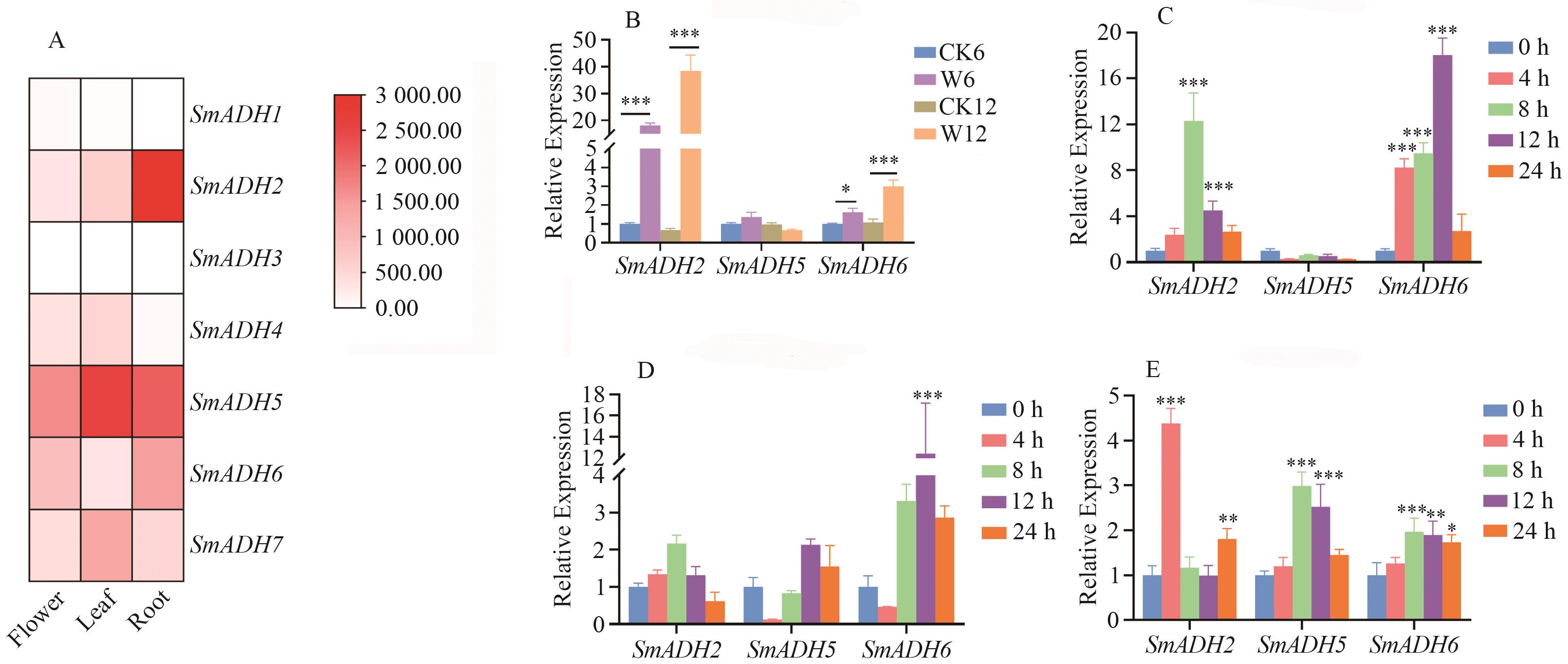
Fig. 5 Expression analysis of S. miltiorrhiza ADHsA: Different tissues; B: waterlogging; C: NaCl;D: GA3;E: PEG-600; *, ** and *** indicate statistically significant differences in gene expression between samples at the 0.05, 0.01, and 0.001 levels, respectively
| [1] | Yang L, Li N, Liu Y, et al. Updates and prospects: morphological, physiological, and molecular regulation in crop response to waterlogging stress [J]. Agronomy, 2023, 13(10): 2599. |
| [2] | Langan P, Bernád V, Walsh J, et al. Phenotyping for waterlogging tolerance in crops: current trends and future prospects [J]. J Exp Bot, 2022, 73(15): 5149-5169. |
| [3] | Ventura I, Brunello L, Iacopino S, et al. Arabidopsis phenotyping reveals the importance of alcohol dehydrogenase and pyruvate decarboxylase for aerobic plant growth [J]. Sci Rep, 2020, 10(1): 16669. |
| [4] | Shen CW, Yuan JP, Ou XQ, et al. Genome-wide identification of alcohol dehydrogenase (ADH) gene family under waterlogging stress in wheat (Triticum aestivum) [J]. PeerJ, 2021, 9: e11861. |
| [5] | Su WH, Ren YJ, Wang DJ, et al. The alcohol dehydrogenase gene family in sugarcane and its involvement in cold stress regulation [J]. BMC Genomics, 2020, 21(1): 521. |
| [6] | Shiao TL, Ellis MH, Dolferus R, et al. Overexpression of alcohol dehydrogenase or pyruvate decarboxylase improves growth of hairy roots at reduced oxygen concentrations [J]. Biotechnol Bioeng, 2002, 77(4): 455-461. |
| [7] | Ismond KP, Dolferus R, de Pauw M, et al. Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway [J]. Plant Physiol, 2003, 132(3): 1292-1302. |
| [8] | Gao MX, Gai CY, Li XY, et al. Waterlogging tolerance of Actinidia Valvata Dunn is associated with high activities of pyruvate decarboxylase, alcohol dehydrogenase and antioxidant enzymes [J]. Plants, 2023, 12(15): 2872. |
| [9] | Tougou M, Hashiguchi A, Yukawa K, et al. Responses to flooding stress in soybean seedlings with the alcohol dehydrogenase transgene [J]. Plant Biotechnol, 2012, 29(3): 301-305. |
| [10] | Wu JJ, Zheng H, Dong YT, et al. The conserved transcriptional regulation mechanism of ADH1 gene in Zanthoxylum armatum to waterlogging stress [J]. Plant Physiol Biochem, 2024, 216: 109133. |
| [11] | Jia QQ, Zhu RY, Tian YM, et al. Salvia miltiorrhiza in diabetes: a review of its pharmacology, phytochemistry, and safety [J]. Phytomedicine, 2019, 58: 152871. |
| [12] | 张玉红, 张玉芳. 泰山丹参生物学特性及其人工繁育技术探讨 [J]. 中国野生植物资源, 2004, 23(3): 64-65. |
| Zhang YH, Zhang YF. Studies on Taishan Danshen biological features and the techniques of artificial breeding [J]. Chin Wild Plant Resour, 2004, 23(3): 64-65. | |
| [13] | Sui C. Salvia miltiorrhiza resources, cultivation, and breeding [M]//The Salvia miltiorrhiza Genome. Cham: Springer International Publishing, 2019: 17-32. |
| [14] | 王振, 李娜, 宋晗, 等. 丹参种植技术研究进展 [J]. 北方园艺, 2023(13): 126-131. |
| Wang Z, Li N, Song H, et al. Research progress on planting technology of Salvia miltiorrhiza [J]. North Hortic, 2023(13): 126-131. | |
| [15] | 路萍, 张利, 王萌, 等. 涝胁迫对不同丹参品系苗期保护酶活性及脂质过氧化作用的影响 [J]. 浙江大学学报: 农业与生命科学版, 2013, 39(1): 56-61. |
| Lu P, Zhang L, Wang M, et al. Effects of water stress on protective enzyme activities and lipid peroxidation in different Salvia miltiorrhiza Bunge varieties at seedling stage [J]. J Zhejiang Univ Agric Life Sci, 2013, 39(1): 56-61. | |
| [16] | Pan X, Chang YJ, Li CL, et al. Chromosome-level genome assembly of Salvia miltiorrhiza with orange roots uncovers the role of Sm2OGD3 in catalyzing 15, 16-dehydrogenation of tanshinones [J]. Hortic Res, 2023, 10(6): uhad069. |
| [17] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [18] | Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era [J]. Mol Biol Evol, 2020, 37(5): 1530-1534. |
| [19] | Zhou H, Jiang MC, Li J, et al. Genome-wide identification and functional analysis of Salvia miltiorrhiza microRNAs reveal the negative regulatory role of smi-miR159a in phenolic acid biosynthesis [J]. Int J Mol Sci, 2024, 25(10): 5148. |
| [20] | Dai XB, Zhuang ZH, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release) [J]. Nucleic Acids Res, 2018, 46(W1): W49-W54. |
| [21] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| [22] | Chen T, Yang M, Cui GH, et al. IMP: bridging the gap for medicinal plant genomics [J]. Nucleic Acids Res, 2024, 52(D1): D1347-D1354. |
| [23] | 化文平, 刘文超, 王喆之, 等. 干涉丹参SmORA1对植物抗病和丹参酮类次生代谢的影响 [J]. 中国农业科学, 2016, 49(3): 491-502. |
| Hua WP, Liu WC, Wang ZZ, et al. Effect of RNAi of SmORA1 on disease resistance and tanshinones secondary metabolism in Salvia miltiorrhiza [J]. Sci Agric Sin, 2016, 49(3): 491-502. | |
| [24] | Xu GX, Guo CC, Shan HY, et al. Divergence of duplicate genes in exon-intron structure [J]. Proc Natl Acad Sci USA, 2012, 109(4): 1187-1192. |
| [25] | Mengarelli DA, Zanor MI. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max) [J]. Planta, 2021, 253(1): 15. |
| [26] | Thompson CE, Freitas LB, Salzano FM. Molecular evolution and functional divergence of alcohol dehydrogenases in animals, fungi and plants [J]. Genet Mol Biol, 2018, 41(1 ): 341-354. |
| [27] | Strommer J. The plant ADH gene family [J]. Plant J, 2011, 66(1): 128-142. |
| [28] | Zhang R, Xuan L, Ni LJ, et al. ADH gene cloning and identification of flooding-responsive genes in Taxodium distichum (L.) rich [J]. Plants, 2023, 12(3): 678. |
| [29] | Xuan L, Hua JF, Zhang F, et al. Identification and functional analysis of ThADH1 and ThADH4 genes involved in tolerance to waterlogging stress in Taxodium hybrid ‘Zhongshanshan 406’ [J]. Genes, 2021, 12(2): 225. |
| [30] | Wang RQ, Du CF, Gu G, et al. Genome-wide identification and expression analysis of the ADH gene family under diverse stresses in tobacco (Nicotiana tabacum L.) [J]. BMC Genomics, 2024, 25(1): 13. |
| [31] | Davik J, Koehler G, From B, et al. Dehydrin, alcohol dehydrogenase, and central metabolite levels are associated with cold tolerance in diploid strawberry (Fragaria spp.) [J]. Planta, 2013, 237(1): 265-277. |
| [1] | REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea [J]. Biotechnology Bulletin, 2025, 41(8): 146-154. |
| [2] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [3] | CHENG Xue, FU Ying, CHAI Xiao-jiao, WANG Hong-yan, DENG Xin. Identification of LHC Gene Family in Setaria italica and Expression Analysis under Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(8): 102-114. |
| [4] | BAI Yu-guo, LI Wan-di, LIANG Jian-ping, SHI Zhi-yong, LU Geng-long, LIU Hong-jun, NIU Jing-ping. Growth-promoting Mechanism of Trichoderma harzianum T9131 on Astragalus membranaceus Seedlings [J]. Biotechnology Bulletin, 2025, 41(8): 175-185. |
| [5] | ZHANG Ze, YANG Xiu-li, NING Dong-xian. Identification of 4CL Gene Family in Arachis hypogaea L. and Expression Analysis in Response to Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(7): 117-127. |
| [6] | LI Xin-ni, LI Jun-yi, MA Xue-hua, HE Wei, LI Jia-li, YU Jia, CAO Xiao-ning, QIAO Zhi-jun, LIU Si-chen. Identification of the PMEI Gene Family of Pectin Methylesterase Inhibitor in Foxtail Millet and Analysis of Its Response to Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(7): 150-163. |
| [7] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [8] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [9] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [10] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [11] | WANG Miao-miao, ZHAO Xiang-long, WANG Zhao-ming, LIU Zhi-peng, YAN Long-feng. Identification of TCP Gene Family in Medicago ruthenica and Their Expression Pattern Analysis under Drought Stress [J]. Biotechnology Bulletin, 2025, 41(6): 179-190. |
| [12] | QU Mei-ling, ZHOU Si-min, ZHANG Jing-yu, HE Jia-wei, ZHU Jia-yuan, LIU Xiao-rong, TONG Qiao-zhen, ZHOU Ri-bao, LIU Xiang-dan. Identification and Expression Analysis of bHLH Transcription Gene Family in Lonicera macranthoides [J]. Biotechnology Bulletin, 2025, 41(6): 256-268. |
| [13] | HUANG Dan, PENG Bing-yang, ZHANG Pan-pan, JIAO Yue, LYU Jia-bin. Identification of HD-Zip Gene Family in Camellia oleifera and Analysis of Its Expression under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(6): 191-207. |
| [14] | LIU Xin, WANG Jia-wen, LI Jin-wei, MOU Ce, YANG Pan-pan, MING Jun, XU Lei-feng. Cloning and Expression Analysis of Three LdBBXs in Lilium davidii var. willmottiae [J]. Biotechnology Bulletin, 2025, 41(5): 186-196. |
| [15] | HE Wei, LI Jun-yi, LI Xin-ni, MA Xue-hua, XING Yuan, CAO Xiao-ning, QIAO Zhi-ju, LIU Si-chen. Genome-wide Identification of U-box E3 Ubiquitin Ligase Gene Family in Setaria italica and Response Analysis to Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(5): 104-118. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||