








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 76-85.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0975
Previous Articles Next Articles
REN Hong-yu1,2( ), PANG Cui-ping1, GU Yang1(
), PANG Cui-ping1, GU Yang1( ), ZHOU Jia-hai1,3(
), ZHOU Jia-hai1,3( )
)
Received:2025-09-11
Online:2026-01-26
Published:2026-02-04
Contact:
GU Yang, ZHOU Jia-hai
E-mail:hy.ren@siat.ac.cn;yang.gu@siat.ac.cn;jiahai@siat.ac.cn
REN Hong-yu, PANG Cui-ping, GU Yang, ZHOU Jia-hai. Multi-strategy Synergy Enhances the Diffraction Resolution of Protein Crystals[J]. Biotechnology Bulletin, 2026, 42(1): 76-85.

Fig. 1 Replacing expression tags and co-expressing molecular chaperones of P450A-C: The expression of P450KstB, P450MciB, and P450ScnB, respectively, with molecular chaperone co-expression and fusion tag swapping. 1: Uninduced strain, 2: SUMO-P450, 3-5: the SUMO-P450 strain was introduced with the molecular chaperones pGro7, pTF16, and pKJE7, respectively, 6: TF-P450, 7: MBP-P450. D: The expression after replacing the GST tag. 1: Uninduced strain, 2-4: P450KstB, P450ScnB, and P450MciB fused with GST tag, respectively
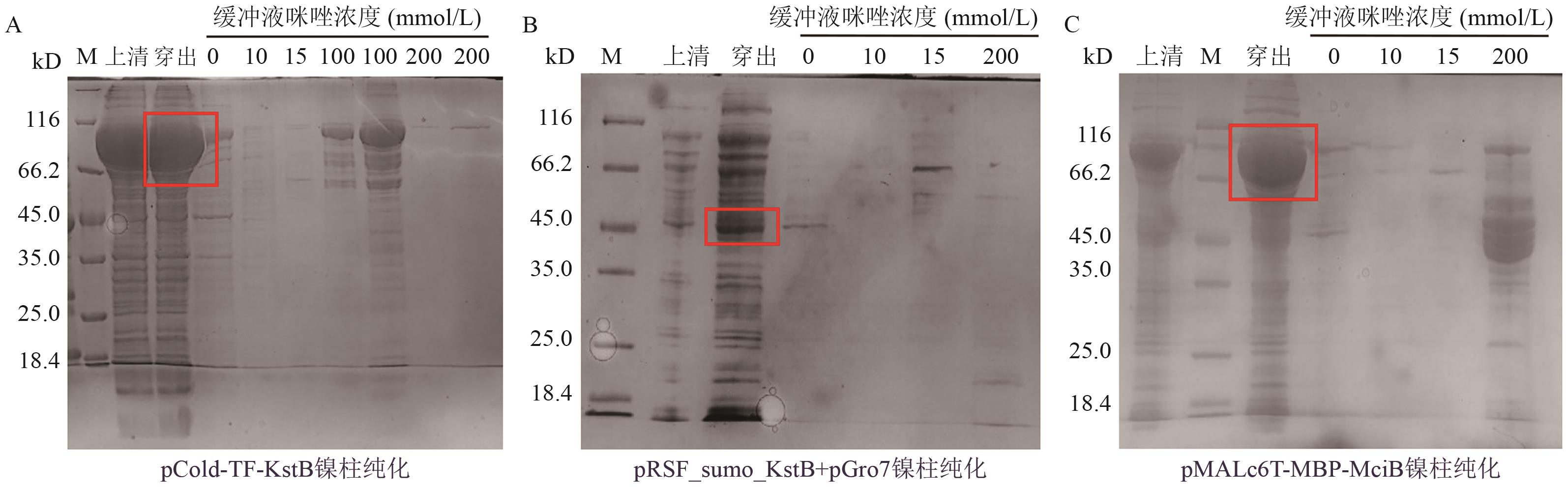
Fig. 2 Ni-NTA analysis of soluble proteins results by Ni-affinity chromatographyA-C: The purification results of affinity chromatography using nickel columns for pCold-TF-KstB, pRSF-sumo-KstB+pGro7, and pMALc6T-MBP-MciB, respectively

Fig. 3 Analysis of P450GpeC protein expressionA: The cell lysate of P450GpeC and the SDS-PAGE analysis of precipitate and supernatant. B: The UV absorption spectrum of P450GpeC with a characteristic absorption peak at 425 nm
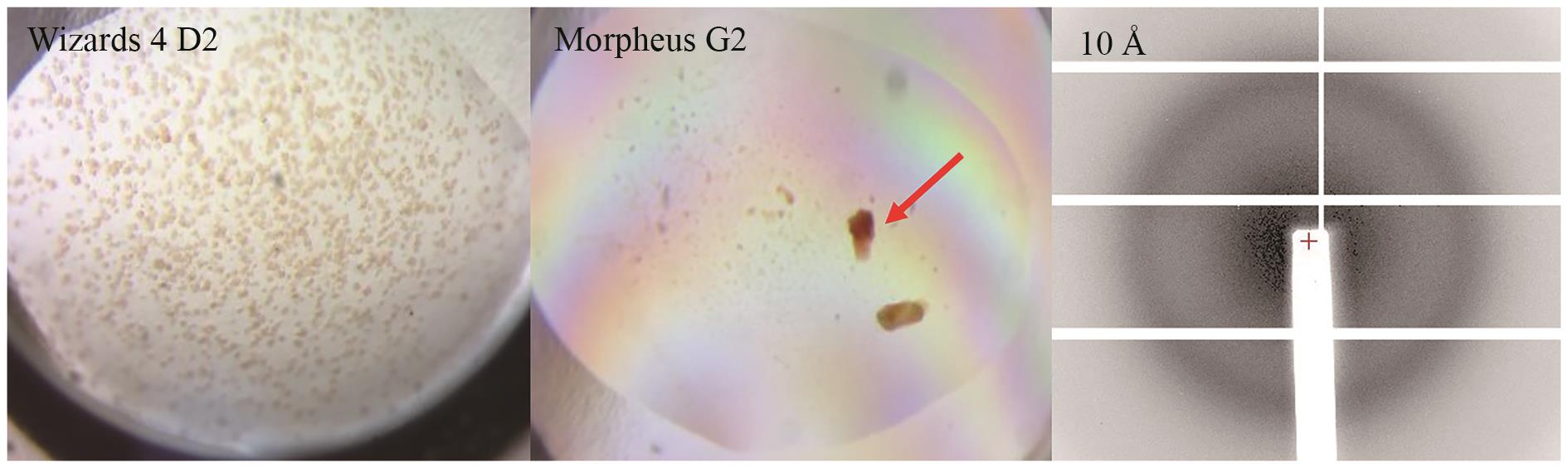
Fig. 7 Crystal images and diffraction patterns of P450GpeC under various initial crystallization conditionsThe arrow indicates the crystal used for diffraction
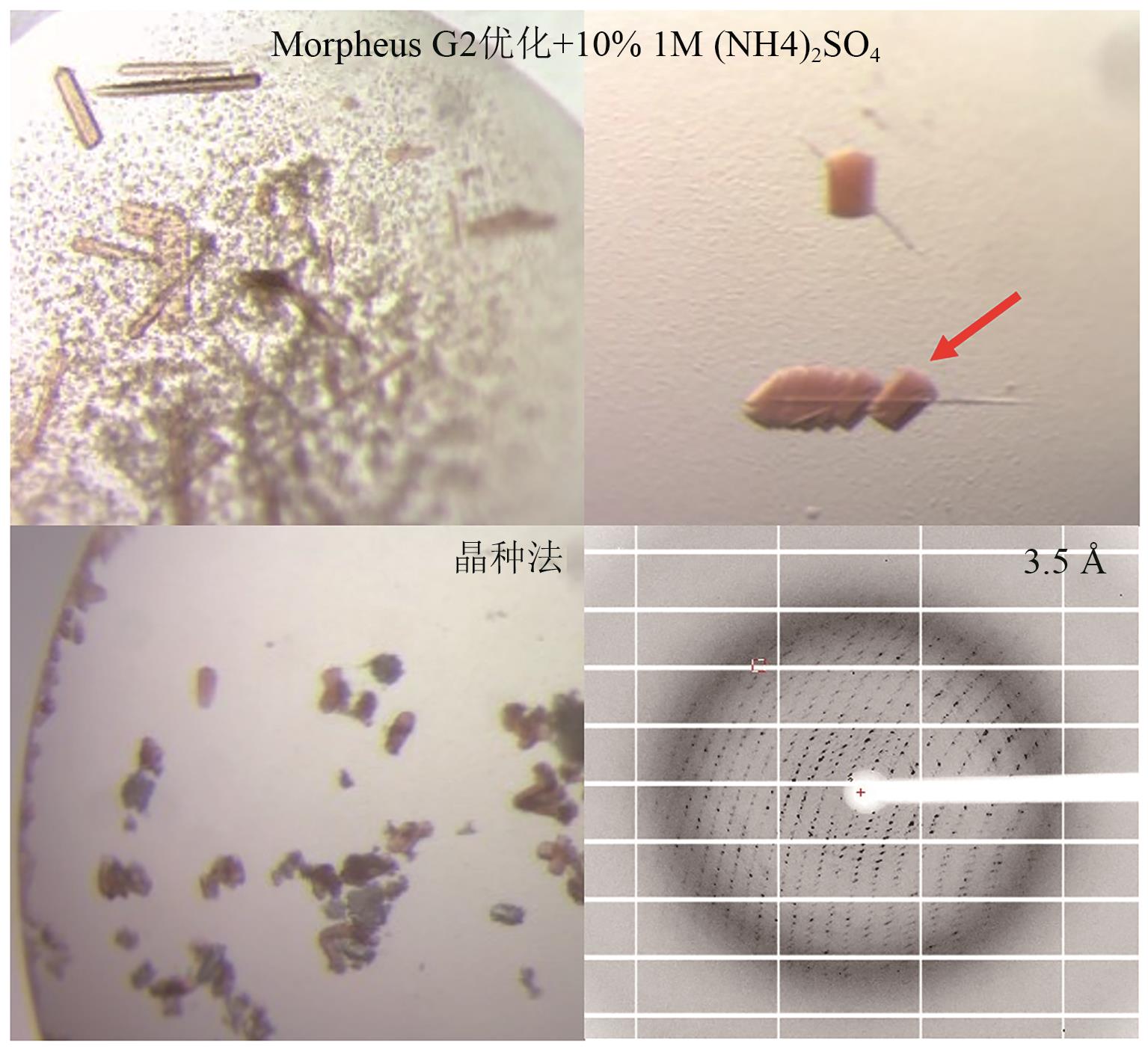
Fig. 8 Crystal images and diffraction patterns of P450GpeC optimized by ammonium sulfate addition and seeding methodThe arrow indicates the crystal used for diffraction
优化方式 Optimization method | 优化效果 Optimization result | 分辨率 Resolution (Å) |
|---|---|---|
| 结晶初筛 | 无法获得蛋白晶体 | - |
| C端loop截短 | 晶体过小或者表面粗糙 | 10 |
| 溶剂优化 | 改善晶体形态 | 5-8 |
| 添加硫酸铵 | 依附盐晶生长 | 3-4 |
| 添加晶种 | 脱离盐晶 | 3-4 |
| 添加SUMO标签 | 外观规则且棱角清晰的高质量集体 | 2.86 |
Table 1 Crystal optimization methods and effects
优化方式 Optimization method | 优化效果 Optimization result | 分辨率 Resolution (Å) |
|---|---|---|
| 结晶初筛 | 无法获得蛋白晶体 | - |
| C端loop截短 | 晶体过小或者表面粗糙 | 10 |
| 溶剂优化 | 改善晶体形态 | 5-8 |
| 添加硫酸铵 | 依附盐晶生长 | 3-4 |
| 添加晶种 | 脱离盐晶 | 3-4 |
| 添加SUMO标签 | 外观规则且棱角清晰的高质量集体 | 2.86 |
| [1] | Papageorgiou AC, Poudel N, Mattsson J. Protein structure analysis and validation with X-ray crystallography [M]//Protein Downstream Processing. New York, NY: Springer US, 2020: 377-404. |
| [2] | Banari A, Samanta AK, Munke A, et al. Advancing time-resolved structural biology: latest strategies in cryo-EM and X-ray crystallography [J]. Nat Meth, 2025, 22(7): 1420-1435. |
| [3] | Nogales E, Mahamid J. Bridging structural and cell biology with cryo-electron microscopy [J]. Nature, 2024, 628(8006): 47-56. |
| [4] | Abramson J, Adler J, Dunger J, et al. Addendum: Accurate structure prediction of biomolecular interactions with AlphaFold 3 [J]. Nature, 2024, 636(8042): E4. |
| [5] | Li FC, Liu RZ, Li WJ, et al. Synchrotron radiation: a key tool for drug discovery [J]. Bioorg Med Chem Lett, 2024, 114: 129990. |
| [6] | Heras B, Edeling MA, Byriel KA, et al. Dehydration converts DsbG crystal diffraction from low to high resolution [J]. Structure, 2003, 11(2): 139-145. |
| [7] | McPherson A, Gavira JA. Introduction to protein crystallization [J]. Acta Crystallogr F Struct Biol Commun, 2014, 70(1): 2-20. |
| [8] | Budziszewski GR, Stojanoff V, Bowman SEJ. Preparing for successful protein crystallization experiments [J]. Acta Crystallogr F Struct Biol Commun, 2025, 81(7): 272-280. |
| [9] | Zhang XF, Xu ZT, Zhou JH, et al. Enhancement of protein crystallization using nano-sized metal-organic framework [J]. Crystals, 2022, 12(5): 578. |
| [10] | Maki S, Hagiwara M. Contactless crystallization method of protein by a magnetic force booster [J]. Sci Rep, 2022, 12: 17287. |
| [11] | Han Q, Brown SJ, Drummond CJ, et al. Protein aggregation and crystallization with ionic liquids: Insights into the influence of solvent properties [J]. J Colloid Interface Sci, 2022, 608: 1173-1190. |
| [12] | Chen TE, Hiramatsu H, Toyouchi S, et al. Laser-polarization-induced anisotropy enhances protein crystallization [J]. Angew Chem Int Ed, 2025, 64(21): e202501827. |
| [13] | Dessau MA, Modis Y. Protein crystallization for X-ray crystallography [J]. JoVE, 2011(47). |
| [14] | Chayen NE, Saridakis E. Protein crystallization: from purified protein to diffraction-quality crystal [J]. Nat Meth, 2008, 5(2): 147-153. |
| [15] | McPherson A, Cudney B. Searching for silver bullets: an alternative strategy for crystallizing macromolecules [J]. J Struct Biol, 2006, 156(3): 387-406. |
| [16] | de Wijn R, Hennig O, Ernst FGM, et al. Combining crystallogenesis methods to produce diffraction-quality crystals of a psychrophilic tRNA-maturation enzyme [J]. Acta Crystallogr F Struct Biol Commun, 2018, 74(11): 747-753. |
| [17] | Khurshid S, Saridakis E, Govada L, et al. Porous nucleating agents for protein crystallization [J]. Nat Protoc, 2014, 9(7): 1621-1633. |
| [18] | Chernov AA. Protein crystals and their growth [J]. J Struct Biol, 2003, 142(1): 3-21. |
| [19] | Zhao MZ, Ma JS, Li M, et al. Cytochrome P450 enzymes and drug metabolism in humans [J]. Int J Mol Sci, 2021, 22(23): 12808. |
| [20] | Sang RY, Jiang WZ, Zhang C, et al. Effect of food components on cytochrome P450 expression and activity [J]. Hum Nutr Metab, 2025, 40: 200304. |
| [21] | Song YR, Li CX, Liu GZ, et al. Drug-metabolizing cytochrome P450 enzymes have multifarious influences on treatment outcomes [J]. Clin Pharmacokinet, 2021, 60(5): 585-601. |
| [22] | Esteves F, Rueff J, Kranendonk M. The central role of cytochrome P450 in xenobiotic metabolism—a brief review on a fascinating enzyme family [J]. J Xenobiot, 2021, 11(3): 94-114. |
| [23] | Greule A, Stok JE, De Voss JJ, et al. Unrivalled diversity: the many roles and reactions of bacterial cytochromes P450 in secondary metabolism 1 [J]. Nat Prod Rep, 2018, 35(8): 757-791. |
| [24] | Rudolf JD, Chang CY, Ma M, et al. Cytochromes P450 for natural product biosynthesis in Streptomyces: sequence, structure, and function [J]. Nat Prod Rep, 2017, 34(9): 1141-1172. |
| [25] | Zhang XW, Guo JW, Cheng FY, et al. Cytochrome P450 enzymes in fungal natural product biosynthesis [J]. Nat Prod Rep, 2021, 38(6): 1072-1099. |
| [26] | He BB, Liu J, Cheng Z, et al. Bacterial cytochrome P450 catalyzed post-translational macrocyclization of ribosomal peptides [J]. Angew Chem Int Ed, 2023, 62(46): e202311533. |
| [27] | Hu YL, Yin FZ, Shi J, et al. P450-modified ribosomally synthesized peptides with aromatic cross-links [J]. J Am Chem Soc, 2023, 145(50): 27325-27335. |
| [28] | Coe LJ, Zhao YW, Padva L, et al. Reassignment of the structure of a tryptophan-containing cyclic tripeptide produced by the biarylitide crosslinking cytochrome P450blt [J]. Chem, 2024, 30(38): e202400988. |
| [29] | Kandy SK, Pasquale MA, Chekan JR. Aromatic side-chain crosslinking in RiPP biosynthesis [J]. Nat Chem Biol, 2025, 21(2): 168-181. |
| [30] | Zdouc MM, Alanjary MM, Zarazúa GS, et al. A biaryl-linked tripeptide from Planomonospora reveals a widespread class of minimal RiPP gene clusters [J]. Cell Chem Biol, 2021, 28(5): 733-739.e4. |
| [31] | Hug JJ, Dastbaz J, Adam S, et al. Biosynthesis of cittilins, unusual ribosomally synthesized and post-translationally modified peptides from Myxococcus xanthus [J]. ACS Chem Biol, 2020, 15(8): 2221-2231. |
| [32] | Wang F, Zhou HY, Olademehin OP, et al. Insights into key interactions between vancomycin and bacterial cell wall structures [J]. ACS Omega, 2018, 3(1): 37-45. |
| [33] | Hu BD, Yu HB, Zhou JW, et al. Whole-cell P450 biocatalysis using engineered Escherichia coli with fine-tuned heme biosynthesis [J]. Adv Sci, 2023, 10(6): 2205580. |
| [34] | Panavas T, Sanders C, Butt TR. SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems [J]. Methods Mol Biol, 2009, 497: 303-317. |
| [1] | WANG Jia, GAO Ming, ZHAO Yun-xiao, CHEN Yi-cun, WANG Yang-dong. Cytochrome P450 Involved in Secondary Metabolites Biosynthesis in Response to Biotic Stresses [J]. Biotechnology Bulletin, 2025, 41(12): 27-39. |
| [2] | TAN Jing-xuan, XING De-xun, HE Tian-jin, LIU Zhan-ying. Advances in Protein Expression System of Pseudomonas fluorescens [J]. Biotechnology Bulletin, 2025, 41(1): 49-61. |
| [3] | PAN Ping-ping, XU Zhi-hao, ZHANG Yi-wen, LI Qing, WANG Zhong-hua. Prokaryotic Expression, Subcellular Localization and Expression Analysis of PcCHS Gene from Polygonatum cyrtonema Hua [J]. Biotechnology Bulletin, 2024, 40(5): 280-289. |
| [4] | ZHANG Yan-feng, YE Li-dan, YU Hong-wei. Redox Partner Engineering: A Solution to the Low Catalytic Efficiency of P450s [J]. Biotechnology Bulletin, 2023, 39(4): 10-23. |
| [5] | QIN Xue-jing, WANG Yu-han, CAO Yi-bo, ZHANG Ling-yun. Prokaryotic Expression and Preparation of Polyclonal Antibody of PwHAP5 Gene in Picea wilsonii [J]. Biotechnology Bulletin, 2022, 38(8): 142-149. |
| [6] | YI Fang, LAI Peng-cheng, ZHENG Xi-ao, HU Shuai, GAO Yan-li. Research on the Preparation and Purification of Kod DNA Polymerase [J]. Biotechnology Bulletin, 2022, 38(5): 183-190. |
| [7] | WEI Qian, LIU Xiao-ning, ZHAO Jie. FoxAl Regulating CYP6B6 Expression Under 2-tridecanone Stress in Helicoverpa armigera [J]. Biotechnology Bulletin, 2022, 38(5): 84-92. |
| [8] | YANG Jia-hui, SUN Yu-ping, LU Ya-ning, LIU huan, LU Cun-fu, CHEN Yu-zhen. Abiotic Stress Resistance of Escherichia coli Transformed with Arabidopsis thaliana AtTERT Gene [J]. Biotechnology Bulletin, 2022, 38(2): 1-9. |
| [9] | CAO Ru-fei, LI Ze-xuan, XU Huan, ZHANG Sha, ZHANG Min-min, DAI Feng, DUAN Xiao-lei. Expression,Purification,and Crystallization of Pif1 Helicase from Bacteroides fragilis [J]. Biotechnology Bulletin, 2021, 37(9): 180-190. |
| [10] | FAN Chen-long, DING Yu. Molecular Cloning and Functional Verification of Histone Deacetylase Gene cobB in Vibrio alginolyticus [J]. Biotechnology Bulletin, 2021, 37(8): 195-202. |
| [11] | TANG Lu, DONG Li-ping, YIN Mo-li, LIU Lei, DONG Yuan, WANG Hui-yan. Preparation and Identification of a Novel FGF20 Monoclonal Antibody [J]. Biotechnology Bulletin, 2021, 37(10): 179-185. |
| [12] | YANG Shi-quan, PENG Dan, FEI Wen-jie, YANG Feng, QU Gao-yi, TANG Wei-wei, OU Jian-ping, DENG Xiang-wen, ZHOU Bo. Cloning and Expression of ClKptA/Tpt1 Gene from Cunninghamia lanceolata(Lamb.)Hook [J]. Biotechnology Bulletin, 2020, 36(8): 15-22. |
| [13] | MIN Qi, GAO Zi-han, YAO Yin, ZHANG Hua-shan, XIONG Hai-rong, ZHANG Li. Effect of Co-expression of HAC1 and Molecular Chaperone Genes on the Expression of Mannanase in Pichia pastoris [J]. Biotechnology Bulletin, 2020, 36(5): 159-168. |
| [14] | MENG Li, DU Cai-ping. Eukaryotic Expression,Purification and Activity Identification of Rat His-Akt1 Recombinant Protein [J]. Biotechnology Bulletin, 2020, 36(12): 98-103. |
| [15] | ZHOU Yang, WANG Tao-tao, YAN Dan-dan, WANG Ying-ying, SHI Qin-xuan, SUN Li-hui, LIN Feng. Advances in Biotechnological Application of Elastin-like Polypeptides as Functional Nanomaterials [J]. Biotechnology Bulletin, 2020, 36(11): 198-208. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||