








生物技术通报 ›› 2021, Vol. 37 ›› Issue (6): 154-162.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1165
收稿日期:2020-09-15
出版日期:2021-06-26
发布日期:2021-07-08
作者简介:张廷焕,男,硕士,助理研究员,研究方向:猪功能基因组;E-mail: 基金资助:
ZHANG Ting-huan( ), ZHANG Li-juan, CHEN Si-qing, GUO Zong-yi(
), ZHANG Li-juan, CHEN Si-qing, GUO Zong-yi( )
)
Received:2020-09-15
Published:2021-06-26
Online:2021-07-08
摘要:
本研究旨在探索miR-378种子序列多态性对其结构、功能和猪胴体性状的影响。利用荣昌猪和亚洲野猪的单核苷酸多态性(SNP)进行群体遗传选择分析,预测miR-378种子序列的多态性对其结构的改变,统计不同猪种中等位基因频率;采用microRNA类似物验证种子序列多态性对miR-378在脂肪细胞中功能以及相关基因表达的影响;运用猪屠宰测定数据与miR-378基因型进行关联分析。结果发现,荣昌猪miR-378位点具有极强的受选择信号,miR-378-3p种子序列第5位出现了A>G的突变,在不同猪种中该位点等位基因频率不同,其中人工选择越强的猪种A等位基因频率越高;该突变会抑制miR-378在脂肪细胞中促进脂质生成的功能,同时会消除miR-378对脂肪分化相关基因(PGC-1α和PGC-1β)以及脂肪分解代谢相关基因(HSL、ATGL和CGI-58)的调控作用;而且该位点的基因型与猪胴体性状显著相关,GG型的背膘厚显著高于AG型和AA型,AA型眼肌面积显著高于GG型。本研究发现的miR-378种子序列这个多态性位点可作为猪生长、胴体性状辅助选择的分子标记应用于猪的育种实践中。
张廷焕, 张利娟, 陈四清, 郭宗义. 猪miR-378种子序列的多态性对其功能以及胴体性状的影响[J]. 生物技术通报, 2021, 37(6): 154-162.
ZHANG Ting-huan, ZHANG Li-juan, CHEN Si-qing, GUO Zong-yi. Effects of the Polymorphism of the Seed Sequence in Porcine miR-378 on Its Function and Carcass Traits[J]. Biotechnology Bulletin, 2021, 37(6): 154-162.
| 引物名称 Name | 引物序列 Sequence | 长度Length/bp |
|---|---|---|
| ap2-F | GGCGTGACTTCCACAAGAGTTTA | 23 |
| ap2-R | GCCTCTTCCTTTGGCTCATG | 20 |
| Ppar-γ-F | GTGAAGGATGCAAGGGTT | 18 |
| Ppar-γ-R | CCTGATGGCATTGTGAGA | 18 |
| C/EBP-α-R | TGGACAAGAACAGCAACGAG | 20 |
| C/EBP-α-F | TCACTGGTCAACTCCAGCAC | 20 |
| PGC-1α-F | AGCGCCGTGTGATTTACGTT | 20 |
| PGC-1α-R | CCGCAGATTTACGGTGCATT | 20 |
| PGC-1β-F | CAGACGTGAGAGCAGAGGGC | 20 |
| PGC-1β-R | CGAATGTATACCACACGGCCT | 21 |
| Resistin-F | CAACTCCCTGTTTCCAAATGC | 21 |
| Resistin-R | GTCCAGCAATTTAAGCCAATGTT | 23 |
| HSL-F | AACCAACCCTAGGCCAACTG | 20 |
| HSL-R | GCTGTGTGCACCAAACTACG | 20 |
| ATGL-F | AGGCCAATGTCTGCAGCACAT | 21 |
| ATGL-R | CAAGTTGTCTGAAATGCCGCC | 21 |
| CGI-58-F | CCCTCAGGTTGGACAAAATGA | 21 |
| CGI-58-R | AGGAAAACCCCATGGCTCTAC | 21 |
| GAPDH-F | ACCACAGTCCATGCCATCAC | 20 |
| GAPDH-R | TCCACCACCCTGTTGCTGTA | 20 |
| U6-F | CGCTTCGGCAGCACATATA | 19 |
| U6-R | TTCACGAATTTGCGTGTCAT | 20 |
| miR-378W | ACTGGACTTGGAGTCAGAAGGC | 22 |
| miR-378M | ACTGGGCTTGGAGTCAGAAGGC | 22 |
表1 RT-PCR引物
Table 1 RT-PCR primers
| 引物名称 Name | 引物序列 Sequence | 长度Length/bp |
|---|---|---|
| ap2-F | GGCGTGACTTCCACAAGAGTTTA | 23 |
| ap2-R | GCCTCTTCCTTTGGCTCATG | 20 |
| Ppar-γ-F | GTGAAGGATGCAAGGGTT | 18 |
| Ppar-γ-R | CCTGATGGCATTGTGAGA | 18 |
| C/EBP-α-R | TGGACAAGAACAGCAACGAG | 20 |
| C/EBP-α-F | TCACTGGTCAACTCCAGCAC | 20 |
| PGC-1α-F | AGCGCCGTGTGATTTACGTT | 20 |
| PGC-1α-R | CCGCAGATTTACGGTGCATT | 20 |
| PGC-1β-F | CAGACGTGAGAGCAGAGGGC | 20 |
| PGC-1β-R | CGAATGTATACCACACGGCCT | 21 |
| Resistin-F | CAACTCCCTGTTTCCAAATGC | 21 |
| Resistin-R | GTCCAGCAATTTAAGCCAATGTT | 23 |
| HSL-F | AACCAACCCTAGGCCAACTG | 20 |
| HSL-R | GCTGTGTGCACCAAACTACG | 20 |
| ATGL-F | AGGCCAATGTCTGCAGCACAT | 21 |
| ATGL-R | CAAGTTGTCTGAAATGCCGCC | 21 |
| CGI-58-F | CCCTCAGGTTGGACAAAATGA | 21 |
| CGI-58-R | AGGAAAACCCCATGGCTCTAC | 21 |
| GAPDH-F | ACCACAGTCCATGCCATCAC | 20 |
| GAPDH-R | TCCACCACCCTGTTGCTGTA | 20 |
| U6-F | CGCTTCGGCAGCACATATA | 19 |
| U6-R | TTCACGAATTTGCGTGTCAT | 20 |
| miR-378W | ACTGGACTTGGAGTCAGAAGGC | 22 |
| miR-378M | ACTGGGCTTGGAGTCAGAAGGC | 22 |
| 引物名称 Name | 引物序列 Sequence | 长度 Length/bp |
|---|---|---|
| miR-378-F | TAACCCCTAGGTGGGTCTGAG | 21 |
| miR-378-R | TCAGATGAGCAGGACAGTTCAG | 22 |
表2 miR-378扩增引物
Table 2 Amplification primer of miR-378
| 引物名称 Name | 引物序列 Sequence | 长度 Length/bp |
|---|---|---|
| miR-378-F | TAACCCCTAGGTGGGTCTGAG | 21 |
| miR-378-R | TCAGATGAGCAGGACAGTTCAG | 22 |
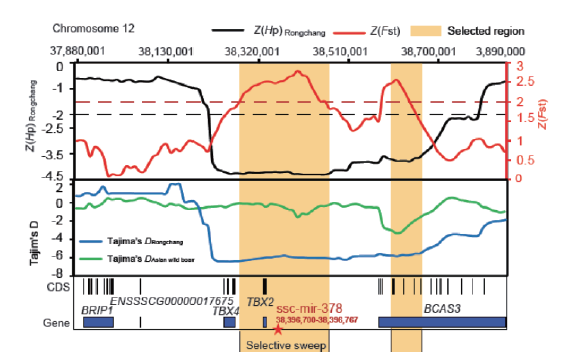
图1 miR-378位点在荣昌猪和亚洲野猪中的群体遗传选择分析 黄色标注的地方表示受选择的区域;红色五角星表示miR-378位点
Fig.1 Population genetic selection analysis of miR-378 locus in Rongchang pig and Asian wild boar The areas marked in yellow indicate the selected regions and the red five-pointed star indicates miR-378 locus
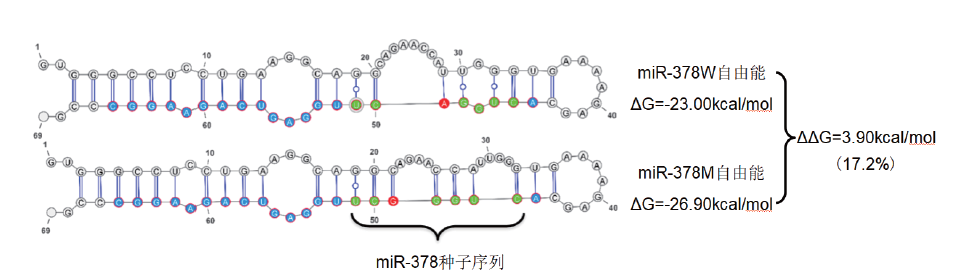
图2 miR-378野生型(miR-378W)和突变型(miR-378M)二级结构以及自由能分析 红色字体表示突变位点
Fig.2 Secondary structure and free energy analysis of miR-378 wild type(miR-378W)and mutated type(miR-378 M) The red font indicates the mutation site
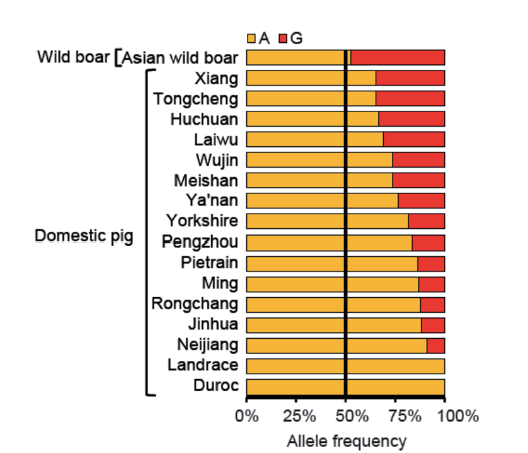
图3 miR-378种子序列第5位碱基(A/G)在不同猪种中的等位基因频率 黄色代表 A 等位基因,红色代表 G 等位基因
Fig.3 Allele frequency of the fifth base(A/G)of miR-378 seed sequence in different pig breeds Yellow represents A allele and red represents G allele
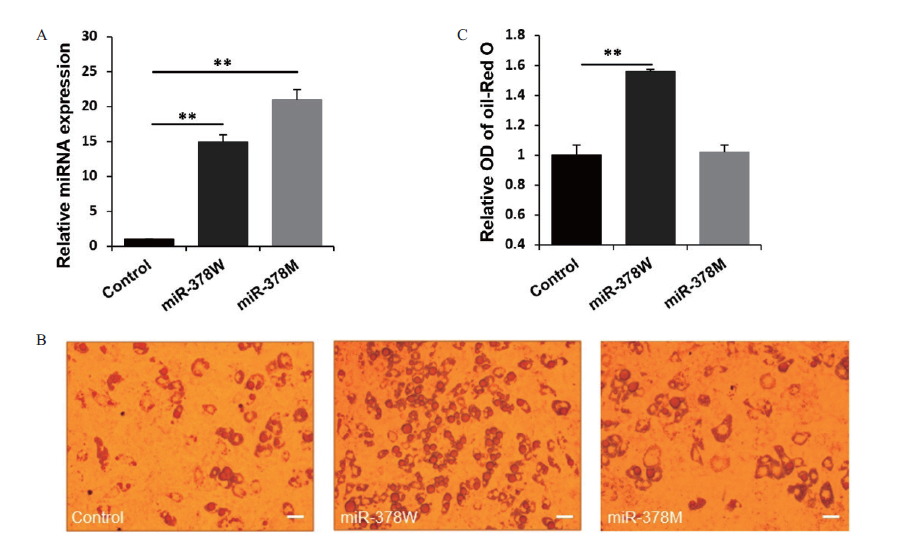
图4 miR-378W和miR-378M对脂肪细胞脂质生成的影响 A:miRNA类似物的转染效率;B:油红 O 染色脂肪细胞,标尺:50 µm;C:定量分析脂质含量。**代表差异显著(P <0.01),下同
Fig.4 Effects of miR-378W and miR-378 M on the lipid production of adipocytes A:Transfection efficiency of miRNA mimics. B:Oil red O staining adipocytes,scale bar:50 μm. C:Quantitative analysis of the lipid content. **refers to significant difference(P < 0.01),the same below
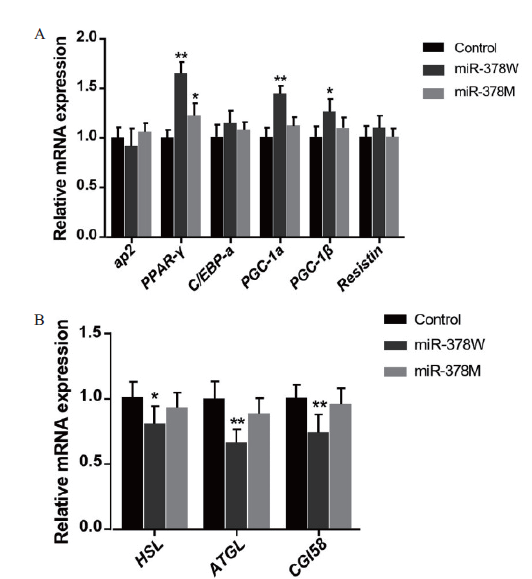
图5 miR-378W和miR-378M对脂质相关基因表达的影响 A:脂肪生成相关基因表达情况;B:脂肪分解相关基因表达情况。*代表差异显著(P <0.05),下同
Fig.5 Effects of miR-378W and miR-378 M on the expres-sion of lipid related genes A:The expression of genes related to adipogenesis. B:The expression of genes related to lipolysis. * refers to significant difference(P < 0.05),the same below

图6 miR-378在10个物种间的碱基序列比对 红色字母代表miR-378的种子序列
Fig.6 Base sequence alignment of miR-378 among 10 species The red letters represent the seed sequence of miR-378
| GG(24) | AG(51) | AA(32) | 加性效应Additive effect | 显性效应Dominant effect | |
|---|---|---|---|---|---|
| 最后肋骨处背膘厚 Backfat thickness at the last rib/cm | 2.77a±0.20 | 2.66b±0.13 | 2.63b±0.16 | 0.07 | -0.04 |
| 眼肌面积 Eye-muscle area/cm2 | 22.16b±1.08 | 23.25ab±0.63 | 23.80a±0.62 | 1.22 | 0.445 |
| 胴体瘦肉率 Lean percentage/% | 48.12±1.06 | 48.28±0.64 | 48.32±0.55 | 0.32 | 0.07 |
表3 miR-378种子序列3种基因型在群体中的个体数及与屠宰性状之间的关联分析
Table 3 Number of individuals for three genotypes of miR-378 seed sequence in the population and their association with slaughter traits
| GG(24) | AG(51) | AA(32) | 加性效应Additive effect | 显性效应Dominant effect | |
|---|---|---|---|---|---|
| 最后肋骨处背膘厚 Backfat thickness at the last rib/cm | 2.77a±0.20 | 2.66b±0.13 | 2.63b±0.16 | 0.07 | -0.04 |
| 眼肌面积 Eye-muscle area/cm2 | 22.16b±1.08 | 23.25ab±0.63 | 23.80a±0.62 | 1.22 | 0.445 |
| 胴体瘦肉率 Lean percentage/% | 48.12±1.06 | 48.28±0.64 | 48.32±0.55 | 0.32 | 0.07 |
| [1] | Yapijakis C. Regulatory role of microRNAs in brain development and function[J]. Adv Exp Med Biol, 2020, 1195:237-247. |
| [2] | Ameres SL, Zamore PD. Diversifying microRNA sequence and function[J]. Nat Rev Mol Cell Biol, 2013, 14(8):475-488. |
| [3] |
Michlewski G, Caceres JF. Post-transcriptional control of miRNA biogenesis[J]. RNA, 2019, 25(1):1-16.
doi: 10.1261/rna.068692.118 pmid: 30333195 |
| [4] | Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions--beyond repression of gene expression[J]. Nat Rev Genet, 2014, 15(9):599-612. |
| [5] |
Dorn GW 2nd, Matkovich SJ, Eschenbacher WH, et al. A human 3’ miR-499 mutation alters cardiac mRNA targeting and function[J]. Circ Res, 2012, 110(7):958-967.
doi: 10.1161/CIRCRESAHA.111.260752 URL |
| [6] |
Kotani A, Ha D, Schotte D, et al. A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells[J]. Cell Cycle, 2010, 9(6):1037-1042.
pmid: 20237425 |
| [7] |
Sun W, Lan J, Chen L, et al. A mutation in porcine pre-miR-15b alters the biogenesis of MiR-15b\16-1 cluster and strand selection of MiR-15b[J]. PLoS One, 2017, 12(5):e0178045.
doi: 10.1371/journal.pone.0178045 URL |
| [8] |
Mencia A, Modamio-Hoybjor S, Redshaw N, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss[J]. Nat Genet, 2009, 41(5):609-613.
doi: 10.1038/ng.355 URL |
| [9] |
Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA[J]. Hum Mol Genet, 2007, 16(9):1124-1131.
doi: 10.1093/hmg/ddm062 URL |
| [10] |
Carrer M, Liu N, Grueter CE, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*[J]. Proc Natl Acad Sci USA, 2012, 109(38):15330-15335.
doi: 10.1073/pnas.1207605109 URL |
| [11] |
Gerin I, Bommer GT, Mccoin CS, et al. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis[J]. Am J Physiol Endocrinol Metab, 2010, 299(2):E198-206.
doi: 10.1152/ajpendo.00179.2010 URL |
| [12] |
Gagan J, Dey BK, Layer R, et al. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation[J]. J Biol Chem, 2011, 286(22):19431-19438.
doi: 10.1074/jbc.M111.219006 URL |
| [13] |
Tong H, Jiang R, Liu T, et al. bta-miR-378 promote the differentiation of bovine skeletal muscle-derived satellite cells[J]. Gene, 2018, 668:246-251.
doi: 10.1016/j.gene.2018.03.102 URL |
| [14] |
Hupkes M, Sotoca AM, Hendriks JM, et al. MicroRNA miR-378 promotes BMP2-induced osteogenic differentiation of mesenchymal progenitor cells[J]. BMC Mol Biol, 2014, 15:1.
doi: 10.1186/1471-2199-15-1 URL |
| [15] |
Chai J, Chen L, Luo Z, et al. Spontaneous single nucleotide polymorphism in porcine microRNA-378 seed region leads to functional alteration[J]. Bioscience, Biotechnology, and Biochemistry, 2018, 82(7):1081-1089.
doi: 10.1080/09168451.2018.1459175 URL |
| [16] |
Xu LL, Shi CM, Xu GF, et al. TNF-alpha, IL-6, and leptin increase the expression of miR-378, an adipogenesis-related microRNA in human adipocytes[J]. Cell Biochem Biophys, 2014, 70(2):771-776.
doi: 10.1007/s12013-014-9980-x URL |
| [17] |
Pan D, Mao C, Quattrochi B, et al. MicroRNA-378 controls classical brown fat expansion to counteract obesity[J]. Nat Commun, 2014, 5:4725.
doi: 10.1038/ncomms5725 URL |
| [18] |
Lu J, Webb R, Richardson JA, et al. MyoR:a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD[J]. Proc Natl Acad Sci USA, 1999, 96(2):552-557.
doi: 10.1073/pnas.96.2.552 URL |
| [19] |
Hou X, Tang Z, Liu H, et al. Discovery of MicroRNAs associated with myogenesis by deep sequencing of serial developmental skeletal muscles in pigs[J]. PLoS One, 2012, 7(12):e52123.
doi: 10.1371/journal.pone.0052123 URL |
| [20] |
Knezevic I, Patel A, Sundaresan NR, et al. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor:implications in postnatal cardiac remodeling and cell survival[J]. J Biol Chem, 2012, 287(16):12913-12926.
doi: 10.1074/jbc.M111.331751 pmid: 22367207 |
| [21] |
Chai J, Chen L, Luo Z, et al. Spontaneous single nucleotide polymorphism in porcine microRNA-378 seed region leads to functional alteration[J]. Biosci Biotechnol Biochem, 2018, 82(7):1081-1089.
doi: 10.1080/09168451.2018.1459175 URL |
| [22] | Wang K, Cao Y, Rong Y, et al. A novel SNP in EIF2AK4 gene is associated with thermal tolerance traits in Chinesecattle[J]. Animals(Basel), 2019, 9(6):375. |
| [23] |
Chang JS, Ghosh S, Newman S, et al. A map of the PGC-1alpha- and NT-PGC-1alpha-regulated transcriptional network in brown adipose tissue[J]. Sci Rep, 2018, 8(1):7876.
doi: 10.1038/s41598-018-26244-4 pmid: 29777200 |
| [24] |
Song W, Zhong C, Yuan Y, et al. Peroxisome proliferator-activated receptor-coactivator 1-beta(PGC-1beta)modulates the expression of genes involved in adipogenesis during preadipocyte differentiation in chicken[J]. Gene, 2020, 741:144516.
doi: 10.1016/j.gene.2020.144516 URL |
| [25] |
Xu L, Ma X, Verma NK, et al. Ablation of PPARgamma in subcutaneous fat exacerbates age-associated obesity and metabolic decline[J]. Aging Cell, 2018, 17(2):e12721.
doi: 10.1111/acel.2018.17.issue-2 URL |
| [26] | Schreiber R, Xie H, Schweiger M. Of mice and men:The physiological role of adipose triglyceride lipase(ATGL)[J]. Biochim Biophys Acta Mol Cell Biol Lipids, 2019, 1864(6):880-899. |
| [27] |
Demine S, Tejerina S, Bihin B, et al. Mild mitochondrial uncoupling induces HSL/ATGL-independent lipolysis relying on a form of autophagy in 3T3-L1 adipocytes[J]. J Cell Physiol, 2018, 233(2):1247-1265.
doi: 10.1002/jcp.25994 URL |
| [28] |
Korbelius M, Vujic N, Sachdev V, et al. ATGL/CGI-58-dependent hydrolysis of a lipid storage pool in murine enterocytes[J]. Cell Rep, 2019, 28(7):1923-1934.e4.
doi: S2211-1247(19)30927-1 pmid: 31412256 |
| [29] |
Choy YH, Park BH, Choi TJ, et al. Estimation of relative economic weights of hanwoo carcass traits based on carcass market price[J]. Asian-Australas J Anim Sci, 2012, 25(12):1667-1673.
doi: 10.5713/ajas.2012.12397 URL |
| [30] |
Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila[J]. EMBO J, 2007, 26(7):1772-1781.
doi: 10.1038/sj.emboj.7601630 URL |
| [31] |
Wang Z, Ma B, Ji X, et al. MicroRNA-378-5p suppresses cell proliferation and induces apoptosis in colorectal cancer cells by targeting BRAF[J]. Cancer Cell Int, 2015, 15:40.
doi: 10.1186/s12935-015-0192-2 URL |
| [32] |
Zeng M, Zhu L, Li L, et al. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1[J]. Cell Mol Biol Lett, 2017, 22:12.
doi: 10.1186/s11658-017-0041-5 URL |
| [33] |
Peng N, Miao Z, Wang L, et al. MiR-378 promotes the cell proliferation of osteosarcoma through down-regulating the expression of Kruppel-like factor 9[J]. Biochem Cell Biol, 2018, 96(5):515-521.
doi: 10.1139/bcb-2017-0186 URL |
| [34] |
Kuang X, Wei C, Zhang T, et al. miR-378 inhibits cell growth and enhances apoptosis in human myelodysplastic syndromes[J]. Int J Oncol, 2016, 49(5):1921-1930.
doi: 10.3892/ijo.2016.3689 URL |
| [35] |
Kulyte A, Lorente-Cebrian S, Gao H, et al. MicroRNA profiling links miR-378 to enhanced adipocyte lipolysis in human cancer cachexia[J]. Am J Physiol Endocrinol Metab, 2014, 306(3):E267-274.
doi: 10.1152/ajpendo.00249.2013 URL |
| [36] |
Li Y, Jiang J, Liu W, et al. microRNA-378 promotes autophagy and inhibits apoptosis in skeletal muscle[J]. Proc Natl Acad Sci USA, 2018, 115(46):E10849-E10858.
doi: 10.1073/pnas.1803377115 URL |
| [1] | 余世洲, 曹领改, 王世泽, 刘勇, 边文杰, 任学良. 烟草种质基因分型核心SNP标记的开发[J]. 生物技术通报, 2023, 39(3): 89-100. |
| [2] | 张廷焕, 郭宗义, 柴捷, 潘红梅, 张亮, 陈磊, 龙熙. 序列变异对miR-378生物发生以及靶标关系的影响[J]. 生物技术通报, 2022, 38(1): 205-214. |
| [3] | 张廷焕, 龙熙, 郭宗义, 柴捷. miR-378促进脂质生成相关靶基因鉴定[J]. 生物技术通报, 2021, 37(2): 80-87. |
| [4] | 郭丽丽, 李昱莹, 郭大龙, 侯小改. 重要花卉植物高密度遗传连锁图谱构建研究进展[J]. 生物技术通报, 2021, 37(1): 246-254. |
| [5] | 张德荣, 马晓霞, 李羽翡, 赵永清, 霍生东, 马忠仁, 柏家林. 脂联素及其受体在哺乳动物中的研究进展与展望[J]. 生物技术通报, 2020, 36(6): 236-244. |
| [6] | 余钧剑, 迟美丽, 贾永义, 刘士力, 竺俊全, 顾志敏. 四引物扩增受阻突变体系PCR技术及其在动植物遗传育种研究中的应用[J]. 生物技术通报, 2020, 36(5): 32-38. |
| [7] | 李晓凯, 范一星, 乔贤, 张磊, 王凤红, 王志英, 王瑞军, 张燕军, 刘志红, 王志新, 何利兵, 李金泉, 苏蕊, 张家新. 山羊基因组与遗传变异图谱研究进展[J]. 生物技术通报, 2020, 36(4): 175-184. |
| [8] | 李标, 张瑞莹, 王小琪, 张存芳, 段子渊. 滩羊微卫星标记多态性及与体尺性状关联分析[J]. 生物技术通报, 2019, 35(6): 131-137. |
| [9] | 黄龙, 吴本丽, 何吉祥, 陈静, 宋光同, 汪翔, 张烨, 武松. 中华鳖MyoD1基因SNP鉴定及其与生长性状的关联分析[J]. 生物技术通报, 2019, 35(4): 76-81. |
| [10] | 王平, 王春语, 张丽霞, 丛玲, 朱振兴, 陆晓春. 利用重测序技术开发高粱多态性SSR分子标记[J]. 生物技术通报, 2019, 35(11): 217-223. |
| [11] | 王燕新, 廖圆圆, 阿依木古丽, 齐骜穹, 李海健, 徐红伟, 杨具田, 蔡勇. 三个绵羊品种LHX3基因多态性与生长性状的关联 分析[J]. 生物技术通报, 2019, 35(10): 162-168. |
| [12] | 纪会, 王会, 柴志欣, 王吉坤, 罗晓林, 姬秋梅, 信金伟, 钟金城. 牦牛miR-378前体克隆及组织表达分析[J]. 生物技术通报, 2019, 35(1): 58-66. |
| [13] | 李晓凯 ,王贵 ,乔贤 ,范一星 ,张磊 ,马宇浩 ,聂瑞雪 ,王瑞军 ,何利兵 ,苏蕊. 全基因组测序在重要家畜上的研究进展[J]. 生物技术通报, 2018, 34(6): 11-21. |
| [14] | 李远,黄洁琼,李佳俊,汪维鹏,张洪建. CYP2C8及CYP3A4细胞表达体系的构建及其在小分子激酶药物对紫衫醇代谢途径抑制研究中的应用[J]. 生物技术通报, 2016, 32(7): 227-233. |
| [15] | 刘娇娇,马友记. 三个绵羊群体MC4R基因多态性及生物信息学分析[J]. 生物技术通报, 2016, 32(4): 87-93. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||