








生物技术通报 ›› 2021, Vol. 37 ›› Issue (11): 285-292.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0101
丰敏1( ), 李舒婷2, 张洋子2, 粟元2, 朱龙佼2, 曹际娟3, 刘海燕1(
), 李舒婷2, 张洋子2, 粟元2, 朱龙佼2, 曹际娟3, 刘海燕1( ), 许文涛2(
), 许文涛2( )
)
收稿日期:2021-01-27
出版日期:2021-11-26
发布日期:2021-12-03
作者简介:丰敏,女,硕士研究生,研究方向:营养与食品卫生学;E-mail: 基金资助:
FENG Min1( ), LI Shu-ting2, ZHANG Yang-zi2, SU Yuan2, ZHU Long-jiao2, CAO Ji-juan3, LIU Hai-yan1(
), LI Shu-ting2, ZHANG Yang-zi2, SU Yuan2, ZHU Long-jiao2, CAO Ji-juan3, LIU Hai-yan1( ), XU Wen-tao2(
), XU Wen-tao2( )
)
Received:2021-01-27
Published:2021-11-26
Online:2021-12-03
摘要:
沙门氏菌是一种常见的食源性致病菌,由沙门氏菌引起的食源性疾病位居榜首。荧光定量聚合酶链式反应(fluorescence quantitative polymerase chain reaction,FQ-PCR)是一种准确、可靠的用于核酸定量的扩增技术,常用的荧光探针法使得扩增体系复杂、引物合成成本高。为了简化FQ-PCR的扩增体系,减少标记基团的修饰,拟通过单标记的荧光自淬灭引物实现新型荧光定量聚合酶链式反应(innovative FQ-PCR,IFQ-PCR)用于模板DNA的定量检测。根据沙门氏菌特异性基因设计单标记发卡型荧光自淬灭引物,并将其应用于FQ-PCR实现了沙门氏菌的检测。设计的自淬灭引物探针一体化,只需单标记,无需额外探针或染料的加入,简化了扩增体系,降低了检测成本;同时引物的发卡结构提高了检测特异性。在最优的引物浓度(0.4 μmol/L)下,沙门氏菌在101-105 CFU/mL的浓度范围内其浓度的对数值与循环阈值(cycle threshold,CT)值之间呈现良好的线性关系,R2高达0.99,检测限低至2 CFU/mL,并且在1.5 h内即可完成扩增检测,方法的稳定性符合要求。因此,一种基于荧光自淬灭引物的IFQ-PCR方法被开发出来并实现了沙门氏菌的简便、快速、灵敏、特异、低成本的检测。
丰敏, 李舒婷, 张洋子, 粟元, 朱龙佼, 曹际娟, 刘海燕, 许文涛. 基于荧光自淬灭引物的沙门氏菌新型荧光定量PCR方法的开发[J]. 生物技术通报, 2021, 37(11): 285-292.
FENG Min, LI Shu-ting, ZHANG Yang-zi, SU Yuan, ZHU Long-jiao, CAO Ji-juan, LIU Hai-yan, XU Wen-tao. Development of a Innovative Fluorescent Quantitative PCR Method for Salmonella Based on Fluorescent Self-quenching Primers[J]. Biotechnology Bulletin, 2021, 37(11): 285-292.
| 名称Name | 序列Sequence(5'-3') | 碱基数Number of bases/nt |
|---|---|---|
| 上游引物Forward primer(FP) | CGGGTCAAGGCTGAGGAA | 18 |
| 原始下游引物Original reverse primer(ORP) | TGCTGAAGTTGAGGATGTTATTCG | 24 |
| 下游引物Reverse primer(RP) 靶基因Target gene | CGAATATGCTGAAGTTGAGGATGTTATT(FAM)CG TGCTGAAGTTGAGGATGTTATTCGCAAAGGGATCCGTCA GACCTCTGGCAGTACCTTCCTCAGCCTTGACCCG | 30 85 |
表1 引物及靶基因序列表
Table 1 Primer and target gene sequence
| 名称Name | 序列Sequence(5'-3') | 碱基数Number of bases/nt |
|---|---|---|
| 上游引物Forward primer(FP) | CGGGTCAAGGCTGAGGAA | 18 |
| 原始下游引物Original reverse primer(ORP) | TGCTGAAGTTGAGGATGTTATTCG | 24 |
| 下游引物Reverse primer(RP) 靶基因Target gene | CGAATATGCTGAAGTTGAGGATGTTATT(FAM)CG TGCTGAAGTTGAGGATGTTATTCGCAAAGGGATCCGTCA GACCTCTGGCAGTACCTTCCTCAGCCTTGACCCG | 30 85 |
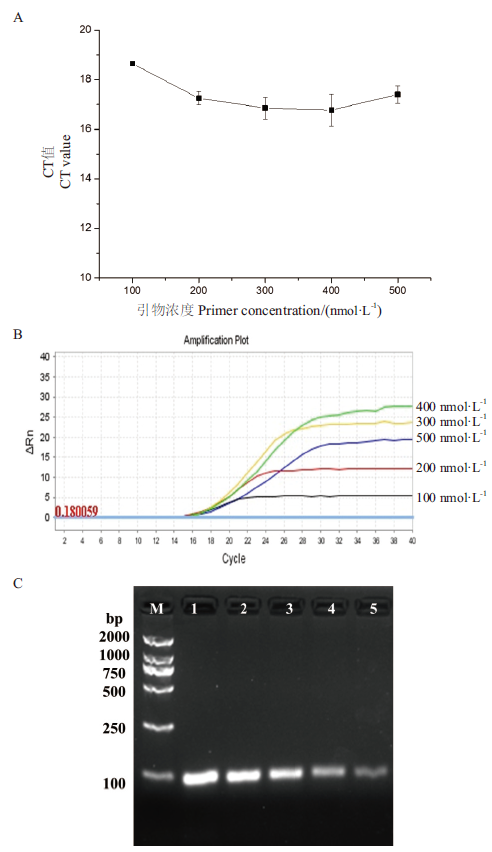
图2 引物浓度优化 A:CT值对比图;B:扩增曲线;C:琼脂糖凝胶电泳图,泳道1-5分别为引物浓度为500 nmol/L、400 nmol/L、300 nmol/L、200 nmol/L、100 nmol/L得到的扩增产物
Fig.2 Primer concentration optimization A: CT value comparison chart. B: Amplification curve. C: Agarose gel electrophoresis chart. Lanes 1-5 are amplified product at primer concentrations of 500 nmol/L, 400 nmol/L, 300 nmol/L, 200 nmol/L, 100 nmol/L, respectively
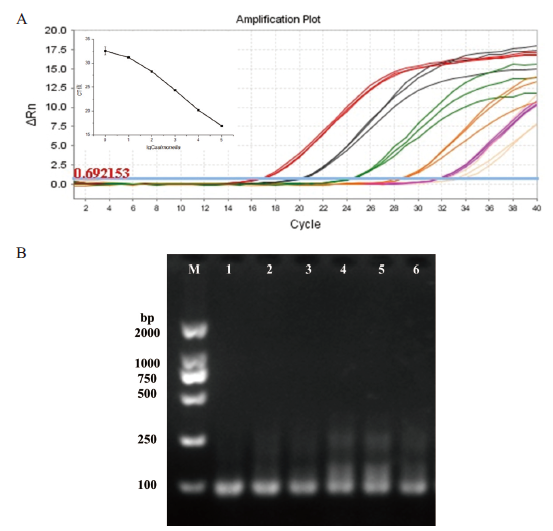
图3 灵敏度检测 A:扩增曲线:大红色线、黑色线、绿色线、橙色线、紫色线、粉红色线对应的沙门氏菌浓度依次为105 CFU/mL、104 CFU/mL、103 CFU/mL、102 CFU/mL、101 CFU/mL、100 CFU/mL(附图为扩增曲线对应的CT值与lgCsalmonella之间的关系图);B:琼脂糖凝胶电泳图:泳道1-6分别对应的沙门氏菌的浓度为105 CFU/mL、104 CFU/mL、103 CFU/mL、102 CFU/mL、101 CFU/mL、100 CFU/mL
Fig.3 Sensitivity detection A: Amplification curve: the Salmonella concentrations corresponding to the big red line, black line, green line, orange line, purple line and pink line are 105 CFU/mL, 104 CFU/mL、103 CFU/mL, 102 CFU/mL, 101 CFU/mL and 100 CFU/mL (the attached shows the relationship between the CT value corresponding to the amplification curve and lgCsalmonella); B: agarose gel electrophoresis diagram: the concentration of Salmonella corresponding to lanes 1-6 is 105 CFU/mL, 104 CFU/mL, 103 CFU/mL, 102 CFU/mL, 101 CFU/mL and 100 CFU/mL
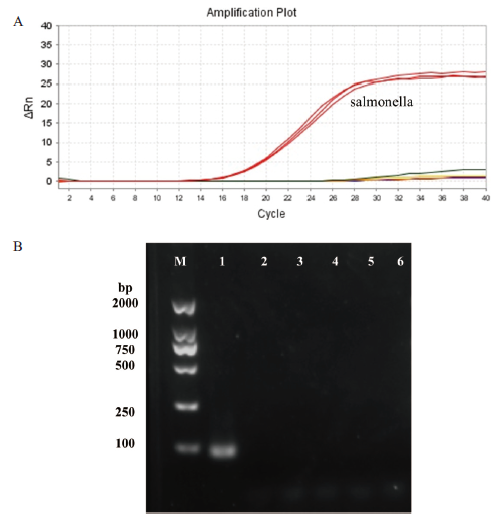
图5 特异性检测 A:扩增曲线:大红色线对应的食源性致病菌为沙门氏菌,其它颜色线分别代表金黄色葡萄球菌、蜡样芽胞杆菌、大肠杆菌、单增李斯特菌、副溶血弧菌;B:琼脂糖凝胶电泳图:泳道1-6分别为沙门氏菌、金黄色葡萄球菌、蜡样芽胞杆菌、大肠杆菌、单增李斯特菌、副溶血弧菌的DNA扩增得到的产物
Fig.5 Specific detection A: Amplification curve: The food-borne pathogen corresponding to the big red line is Salmonella, and the other color lines represent Staphylococcus aureus, Bacillus cereus, Escherichia coli, Listeria monocytogenes, and Vibrio parahaemolyticus. B :Agarose gel electrophoresis diagram: lanes 1-6 are the products of DNA amplification of Salmonella, Staphylococcus aureus, Bacillus cereus, Escherichia coli, Listeria monocytogenes, and Vibrio parahaemolyticus, respectively
| 沙门氏菌的浓度 Concentration of Salmonella/(CFU·mL-1) | CT值 CT value | ±S | RSD | ||
|---|---|---|---|---|---|
| 105 | 16.9 | 16.98 | 16.89 | 16.9±0.05 | 0.29% |
| 104 | 20.26 | 20.29 | 20.18 | 20.2±0.05 | 0.28% |
| 103 | 23.66 | 23.63 | 23.55 | 23.6±0.06 | 0.24% |
| 102 | 26.49 | 26.43 | 26.34 | 26.4±0.08 | 0.29% |
表2 IFQ-PCR方法对牛奶样品中不同浓度的沙门氏菌的检测结果
Table 2 IFQ-PCR method for the detection results of different concentrations of Salmonella in milk samples
| 沙门氏菌的浓度 Concentration of Salmonella/(CFU·mL-1) | CT值 CT value | ±S | RSD | ||
|---|---|---|---|---|---|
| 105 | 16.9 | 16.98 | 16.89 | 16.9±0.05 | 0.29% |
| 104 | 20.26 | 20.29 | 20.18 | 20.2±0.05 | 0.28% |
| 103 | 23.66 | 23.63 | 23.55 | 23.6±0.06 | 0.24% |
| 102 | 26.49 | 26.43 | 26.34 | 26.4±0.08 | 0.29% |
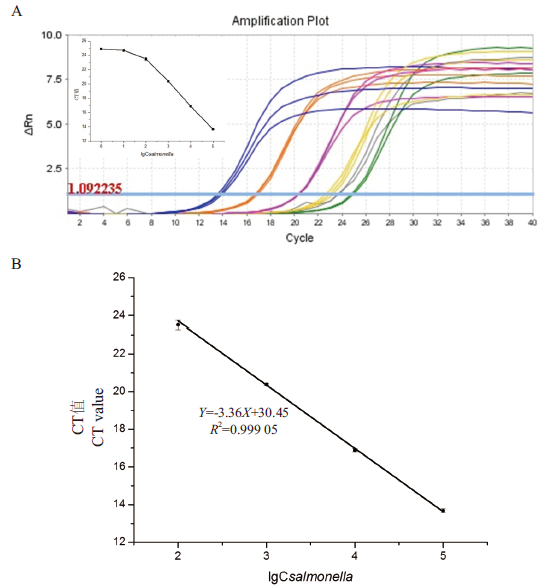
图7 基于染料法的FQ-PCR的灵敏度检测 A:扩增曲线:蓝色线、橙色线、紫色线、黄色线、灰色线、绿色线对应的沙门氏菌浓度依次为105 CFU/mL、104 CFU/mL、103 CFU/mL、102 CFU/mL、101 CFU/mL、100 CFU/mL(附图为扩增曲线对应的CT值与lgCsalmonella之间的关系图);B:FQ-PCR法标准曲线图
Fig.7 Sensitivity detection of FQ-PCR based on dye method A: Amplification curve: The Salmonella concentrations corresponding to the blue line, orange line, purple line, yellow line, gray line, and green line are 105 CFU/mL, 104 CFU/mL, 103 CFU/mL, 102 CFU/mL, 101 CFU/mL and 100 CFU/mL (The attached shows the relationship between the CT value corresponding to the amplification curve and lgCsalmonella). B: FQ-PCR method standard curve
| 方法Method | 优点Advantage | 缺点Disadvantage |
|---|---|---|
| IFQ-PCR法 IFQ-PCR method | 引物的修饰简单、合成成本相对较低、扩增体系简单、可减少引物二聚体和碱基错配的发生 | 引物的设计要求高 |
| FQ-PCR染料法 FQ-PCR dye method | 无需标记、灵敏度高、成本低、可通过熔解曲线分析结果 | 特异性差、扩增体系需额外添加染料 |
| 探针法 Probe method | 特异性最强、不受非特异性扩增与引物二聚体的影响 | 探针的设计复杂、标记成本高、扩增体系复杂 |
表3 IFQ-PCR方法与FQ-PCR方法的比较[13-16,19-23]
Table 3 Comparison of IFQ-PCR method and FQ-PCR method [13-16, 19-23]
| 方法Method | 优点Advantage | 缺点Disadvantage |
|---|---|---|
| IFQ-PCR法 IFQ-PCR method | 引物的修饰简单、合成成本相对较低、扩增体系简单、可减少引物二聚体和碱基错配的发生 | 引物的设计要求高 |
| FQ-PCR染料法 FQ-PCR dye method | 无需标记、灵敏度高、成本低、可通过熔解曲线分析结果 | 特异性差、扩增体系需额外添加染料 |
| 探针法 Probe method | 特异性最强、不受非特异性扩增与引物二聚体的影响 | 探针的设计复杂、标记成本高、扩增体系复杂 |
| [1] |
Hao LL, Gu HJ, et al. A chemiluminescent aptasensor based on rolling circle amplification and Co2+/N-(aminobutyl)-N-(ethylisoluminol)functional flowerlike gold nanoparticles for Salmonella typhimurium detection[J]. Talanta, 2017, 164: 275-282.
doi: 10.1016/j.talanta.2016.11.053 URL |
| [2] |
Scharff RL. Economic burden from health losses due to foodborne illness in the United States[J]. J Food Prot, 2012, 75(1): 123-131.
doi: 10.4315/0362-028X.JFP-11-058 URL |
| [3] |
Wang L, et al. QCM-based aptamer selection and detection of Salmonella typhimurium[J]. Food Chem, 2017, 221: 776-782.
doi: 10.1016/j.foodchem.2016.11.104 URL |
| [4] |
Li ST, et al. Luminescent DNAzyme and universal blocking linker super polymerase chain reaction visual biosensor for the detection of Salmonella[J]. Food Chem, 2020, 324: 126859.
doi: 10.1016/j.foodchem.2020.126859 URL |
| [5] |
Paniel N, Baudart J, Hayat A, et al. Aptasensor and genosensor methods for detection of microbes in real world samples[J]. Methods, 2013, 64(3): 229-240.
doi: 10.1016/j.ymeth.2013.07.001 pmid: 23872322 |
| [6] |
Narmani A, Rezvani M, Farhood B, et al. Folic acid functionalized nanoparticles as pharmaceutical carriers in drug delivery systems[J]. Drug Dev Res, 2019, 80(4): 404-424.
doi: 10.1002/ddr.v80.4 URL |
| [7] | 孙晓飞, 赵凯, 王金斌, 等. 食源性病原菌检测方法研究进展[J]. 现代农业科技, 2010(14): 334-336. |
| Sun XF, Zhao K, Wang JB, et al. Research progress of detection technology in foodborne pathogens[J]. Mod Agric Sci Technol, 2010(14): 334-336. | |
| [8] | 张冲, 刘祥, 陈计峦. 实时荧光定量RT-PCR检测沙门氏菌活菌[J]. 食品工业科技, 2012, 33(6): 91-94. |
| Zhang C, Liu X, Chen JL. Real-time quantitative reverse transcription polymerase chain reaction detection of live Salmonella[J]. Sci Technol Food Ind, 2012, 33(6): 91-94. | |
| [9] | 王凤军, 叶素丹. TaqMan实时荧光PCR对沙门氏菌能力验证样品的快速检测与鉴定[J]. 现代食品科技, 2020, 36(12): 300-306, 83. |
| Wang FJ, Ye SD. Rapid detection and identification of Salmonella in proficiency testing samples by TaqMan real-time fluorescent PCR[J]. Mod Food Sci Technol, 2020, 36(12): 300-306, 83. | |
| [10] | Singh LP, Yadav AS, Singh RP, et al. Detection and quantification of Salmonella on chicken egg shell by real time PCR[J]. Indian J Animal Sci, 2013, 83(5): 481-483. |
| [11] |
Lee N, Kwon KY, Oh SK, et al. A multiplex pcr assay for simultaneous detection of Escherichia coli O157:H7, Bacillus cereus, Vibrio parahaemolyticus, Salmonella spp. Listeria monocytogenes, and Staphylococcus aureus in Korean Ready-to-Eat food[J]. Foodborne Pathog Dis, 2014, 11(7): 574-580.
doi: 10.1089/fpd.2013.1638 URL |
| [12] |
Rodríguez A, Rodríguez M, Córdoba JJ, et al. Design of primers and probes for quantitative real-time PCR methods[J]. Methods Mol Biol, 2015, 1275: 31-56.
doi: 10.1007/978-1-4939-2365-6_3 pmid: 25697650 |
| [13] |
Nazarenko I, Lowe B, Darfler M, et al. Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore[J]. Nucleic Acids Res, 2002, 30(9): e37.
doi: 10.1093/nar/30.9.e37 URL |
| [14] |
Crockett AO, Wittwer CT. Fluorescein-labeled oligonucleotides for real-time pcr:using the inherent quenching of deoxyguanosine nucleotides[J]. Anal Biochem, 2001, 290(1): 89-97.
pmid: 11180941 |
| [15] |
Nazarenko I, et al. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes[J]. Nucleic Acids Res, 2002, 30(9): 2089-2195.
pmid: 11972350 |
| [16] |
Knemeyer JP, Marmé N, Sauer M. Probes for detection of specific DNA sequences at the single-molecule level[J]. Anal Chem, 2000, 72(16): 3717-3724.
pmid: 10959954 |
| [17] |
Zhou ZQ, Zhang YZ, Guo MZ, et al. Ultrasensitive magnetic DNAzyme-copper nanoclusters fluorescent biosensor with triple amplification for the visual detection of E. coli O157:H7[J]. Biosens Bioelectron, 2020, 167: 112475.
doi: 10.1016/j.bios.2020.112475 URL |
| [18] |
Hui CY, Liu M, Li YF, et al. A paper sensor printed with multifunctional bio/nano materials[J]. Angew Chem Int Ed, 2018, 57(17): 4549-4553.
doi: 10.1002/anie.v57.17 URL |
| [19] |
Rani N, Vajpayee P, Bhatti S, et al. Quantification of Salmonella Typhi in water and sediments by molecular-beacon based qPCR[J]. Ecotoxicol Environ Saf, 2014, 108: 58-64.
doi: 10.1016/j.ecoenv.2014.06.033 URL |
| [20] |
Li B, Chen JQ. Development of a sensitive and specific qPCR assay in conjunction with propidium monoazide for enhanced detection of live Salmonella spp. in food[J]. BMC Microbiol, 2013, 13: 273.
doi: 10.1186/1471-2180-13-273 URL |
| [21] |
Kim SA, Park SH, Lee SI, et al. Development of a rapid method to quantify Salmonella Typhimurium using a combination of MPN with qPCR and a shortened time incubation[J]. Food Microbiol, 2017, 65: 7-18.
doi: 10.1016/j.fm.2017.01.013 URL |
| [22] |
Richards AK, Hopkins BA, Shariat NW. Conserved CRISPR arrays in Salmonella enterica serovar Infantis can serve as qPCR targets to detect Infantis in mixed serovar populations[J]. Lett Appl Microbiol, 2020, 71(2): 138-145.
doi: 10.1111/lam.v71.2 URL |
| [23] |
Azinheiro S, Carvalho J, Prado M, et al. Multiplex detection of Salmonella spp. E. coli O157 and L. monocytogenes by qPCR melt curve analysis in spiked infant formula[J]. Microorganisms, 2020, 8(9): 1359.
doi: 10.3390/microorganisms8091359 URL |
| [1] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [2] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [3] | 宋海娜, 吴心桐, 杨鲁豫, 耿喜宁, 张华敏, 宋小龙. 葱鳞葡萄孢菌诱导下韭菜RT-qPCR内参基因的筛选和验证[J]. 生物技术通报, 2023, 39(3): 101-115. |
| [4] | 穆德添, 万凌云, 章瑶, 韦树根, 陆英, 付金娥, 田艺, 潘丽梅, 唐其. 钩藤管家基因筛选及生物碱合成相关基因的表达分析[J]. 生物技术通报, 2023, 39(2): 126-138. |
| [5] | 程深伟, 张克强, 梁军锋, 刘福元, 郜兴亮, 杜连柱. 畜禽养殖粪污中典型致病菌的三重微滴式数字PCR定量检测方法的建立[J]. 生物技术通报, 2022, 38(9): 271-280. |
| [6] | 刘理慧, 储锦华, 隋雨欣, 陈杨, 程古月. 沙门氏菌中主要毒力因子的研究进展[J]. 生物技术通报, 2022, 38(9): 72-83. |
| [7] | 曹英芳, 赵新, 刘双, 李瑞环, 刘娜, 徐石勇, 高芳瑞, 马卉, 兰青阔, 檀建新, 王永. 抗除草剂大豆GE-J12实时荧光定量PCR检测方法的建立[J]. 生物技术通报, 2022, 38(7): 146-152. |
| [8] | 王子琰, 王建, 张伦, 桂水清, 卢雪梅. 家蝇抗菌肽MDC对鼠伤寒沙门氏菌的抑菌稳定性研究[J]. 生物技术通报, 2022, 38(3): 149-156. |
| [9] | 粟元, 朱龙佼, 曹继娟, 刘建龙, 许文涛. 基于大肠埃希菌 O157∶H7的荧光定量冻干检测试剂盒的研制[J]. 生物技术通报, 2022, 38(3): 264-275. |
| [10] | 兰欣悦, 刘宁宁, 朱龙佼, 陈旭, 褚华硕, 李相阳, 段诺, 许文涛. 四环素双价适配体非酶免标记传感器[J]. 生物技术通报, 2022, 38(3): 276-284. |
| [11] | 蒋旭东, 刘宇, 邬建飞, 胡双阁, 卢建远, 字向东. 牦牛FGG组织表达与雌性生殖器官中定位分析[J]. 生物技术通报, 2022, 38(11): 286-294. |
| [12] | 徐圆圆, 赵国春, 郝颖颖, 翁学煌, 陈仲, 贾黎明. 无患子RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2022, 38(10): 80-89. |
| [13] | 陈建军, 赵怡迪, 曹香林. 脂多糖对鲤肠上皮细胞转录组模式的调控分析[J]. 生物技术通报, 2021, 37(8): 213-220. |
| [14] | 黄景晓, 尚俊康, 陈慧敏, 沈嘉旻, 黎圆圆, 喻玉立, 倪进东, 林伯坤. 一株烈性沙门氏菌噬菌体的生物学特性及基因组分析[J]. 生物技术通报, 2021, 37(6): 136-146. |
| [15] | 王欢禹, 常昊宛, 章崇祺, 金卫林, 魏芳. 五种检测嵌合抗原受体表达方法的比较[J]. 生物技术通报, 2021, 37(12): 265-273. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||