








生物技术通报 ›› 2022, Vol. 38 ›› Issue (8): 77-83.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1368
蔡佳1,2( ), 梁振宇1, 黄瑜1, 鲁义善1, 施钢1, 简纪常1
), 梁振宇1, 黄瑜1, 鲁义善1, 施钢1, 简纪常1
收稿日期:2021-10-29
出版日期:2022-08-26
发布日期:2022-09-14
作者简介:蔡佳,男,博士,副教授,研究方向:鱼类免疫学、水生动物病毒致病机理;E-mail: 基金资助:
CAI Jia1,2( ), LIANG Zhen-yu1, HUANG Yu1, LU Yi-shan1, SHI gang1, JIAN Ji-chang1
), LIANG Zhen-yu1, HUANG Yu1, LU Yi-shan1, SHI gang1, JIAN Ji-chang1
Received:2021-10-29
Published:2022-08-26
Online:2022-09-14
摘要:
以参与石斑鱼抗病毒免疫反应的EcBAG3蛋白为研究对象,采用酵母双杂交等技术筛选与鉴定EcBAG3的互作蛋白。首先构建了石斑鱼脾脏细胞的酵母双杂交cDNA文库,文库的容量为5.6×106 CFU,插入片段的平均长度在750 bp以上,重组率为100%。进一步利用此文库筛选EcBAG3的互作因子,初步得到了83个阳性克隆。对这些克隆进行测序与一对一互作验证,最终获得7个候选互作蛋白,功能预测结果显示,互作蛋白涉及免疫调控、转录调节、胁迫响应、信号转导等方面的功能,为进一步解析石斑鱼BAG3在病毒感染中的作用机制奠定了基础。
蔡佳, 梁振宇, 黄瑜, 鲁义善, 施钢, 简纪常. 利用酵母双杂交系统筛选与鉴定石斑鱼EcBAG3互作因子[J]. 生物技术通报, 2022, 38(8): 77-83.
CAI Jia, LIANG Zhen-yu, HUANG Yu, LU Yi-shan, SHI gang, JIAN Ji-chang. Screening and Identifing the Interacting Proteins of Grouper(Epinephelus coioides)EcBAG3 Using Yeast Two-hybrid System[J]. Biotechnology Bulletin, 2022, 38(8): 77-83.
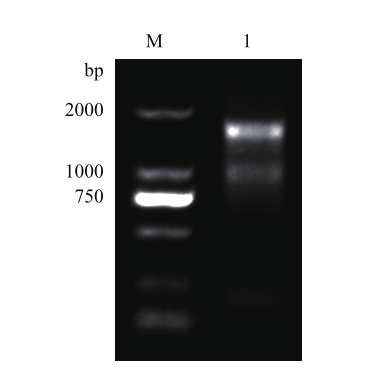
图1 总RNA琼脂糖凝胶检测 M:DL2000 DNA marker;1:GS细胞提取的总RNA
Fig. 1 Agarose gel electrophoresis for the detection of total RNA M:DL2000 DNA marker. 1:Total RNA extracted from GS cells
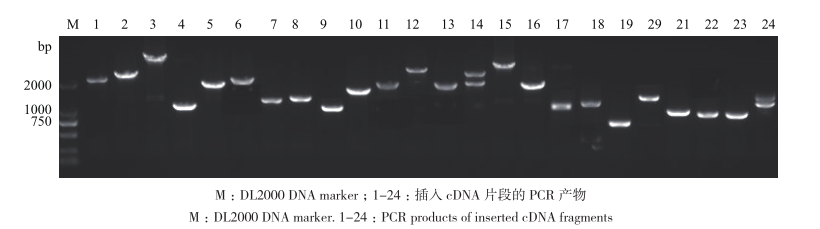
图2 文库部分插入片段琼脂糖凝胶检测 M:DL2000 DNA marker;1-24:插入cDNA片段的PCR 产物
Fig. 2 Agarose gel electrophoresis for the detection of partial inserted cDNA fragments in library M:DL2000 DNA marker. 1-24:PCR products of inserted cDNA fragments
| 候选蛋白 Candidate protein | NCBI登录号 Accession number of NCBI |
|---|---|
| Heat Shock Cognate 71 kD Protein(Hsc71) | XP_033491860.1 |
| Heat Shock Protein 70(Hsp70) | AWD76391.1 |
| Filamin-A | XP_033484911.1 |
| Collagen,Type V,Alpha 2a | XP_033473099.1 |
| Zinc Finger Protein 217(ZNF217) | XP_033484552.1 |
| CCN Family Member 1(CCN1) | XP_033476535.1 |
| Suppressor Of Cytokine Signaling 9(SOCS9) | XP_033486547.1 |
表1 酵母双杂交阳性克隆比对结果
Table 1 Alignments of positive clones screened through yeast two hybrid
| 候选蛋白 Candidate protein | NCBI登录号 Accession number of NCBI |
|---|---|
| Heat Shock Cognate 71 kD Protein(Hsc71) | XP_033491860.1 |
| Heat Shock Protein 70(Hsp70) | AWD76391.1 |
| Filamin-A | XP_033484911.1 |
| Collagen,Type V,Alpha 2a | XP_033473099.1 |
| Zinc Finger Protein 217(ZNF217) | XP_033484552.1 |
| CCN Family Member 1(CCN1) | XP_033476535.1 |
| Suppressor Of Cytokine Signaling 9(SOCS9) | XP_033486547.1 |
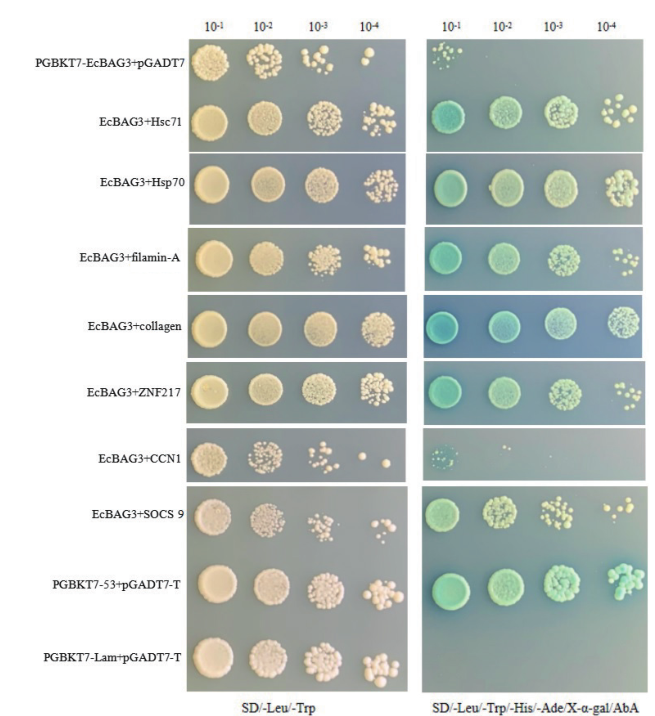
图5 EcBAG3与候选互作蛋白验证 阳性对照:pGBKT7-53+pGADT7-T;阴性对照:pGBKT7-lam+pGADT7;阴性对照:pGBKT7-EcBAG3+pGADT7
Fig. 5 Verification of EcBAG3 and candidate interacting protein Positive control:pGBKT7-53+pGADT7-T. Negative control:pGBKT7-lam+pGADT7. Negative control:pGBKT7-EcBAG3+pGADT7
| [1] |
Qin QW, Chang SF, Ngoh-Lim GH, et al. Characterization of a novel Ranavirus isolated from grouper Epinephelus tauvina[J]. Dis Aquat Org, 2003, 53:1-9.
doi: 10.3354/dao053001 URL |
| [2] |
Liu H, Teng Y, Zheng XC, et al. Complete sequence of a viral nervous necrosis virus(NNV)isolated from red-spotted grouper(Epinephelus akaara)in China[J]. Arch Virol, 2012, 157(4):777-782.
doi: 10.1007/s00705-011-1187-5 URL |
| [3] |
Huang XH, Huang YH, Ouyang ZL, et al. Singapore grouper Iridovirus, a large DNA virus, induces nonapoptotic cell death by a cell type dependent fashion and evokes ERK signaling[J]. Apoptosis, 2011, 16(8):831-845.
doi: 10.1007/s10495-011-0616-y URL |
| [4] |
Li C, Wang LQ, Liu JX, et al. Singapore grouper Iridovirus(SGIV)inhibited autophagy for efficient viral replication[J]. Front Microbiol, 2020, 11:1446.
doi: 10.3389/fmicb.2020.01446 URL |
| [5] |
Huang YH, Zhang Y, Liu ZT, et al. Autophagy participates in lysosomal vacuolation-mediated cell death in RGNNV-infected cells[J]. Front Microbiol, 2020, 11:790.
doi: 10.3389/fmicb.2020.00790 URL |
| [6] |
Li C, Liu JX, Zhang X, et al. Red grouper nervous necrosis virus(RGNNV)induces autophagy to promote viral replication[J]. Fish Shellfish Immunol, 2020, 98:908-916.
doi: 10.1016/j.fsi.2019.11.053 URL |
| [7] |
Huang YH, Huang XH, Yang Y, et al. Involvement of fish signal transducer and activator of transcription 3(STAT3)in nodavirus infection induced cell death[J]. Fish Shellfish Immunol, 2015, 43(1):241-248.
doi: 10.1016/j.fsi.2014.12.031 URL |
| [8] |
Bruno AP, De Simone FI, Iorio V, et al. HIV-1 Tat protein induces glial cell autophagy through enhancement of BAG3 protein levels[J]. Cell Cycle, 2014, 13(23):3640-3644.
doi: 10.4161/15384101.2014.952959 URL |
| [9] |
Rosati A, Graziano V, De Laurenzi V, et al. BAG3:a multifaceted protein that regulates major cell pathways[J]. Cell Death Dis, 2011, 2:e141.
doi: 10.1038/cddis.2011.24 URL |
| [10] |
Shi HY, Xu HD, Li ZJ, et al. BAG3 regulates cell proliferation, migration, and invasion in human colorectal cancer[J]. Tumour Biol, 2016, 37(4):5591-5597.
doi: 10.1007/s13277-015-4403-1 URL |
| [11] |
Behl C. Breaking BAG:the co-chaperone BAG3 in health and disease[J]. Trends Pharmacol Sci, 2016, 37(8):672-688.
doi: 10.1016/j.tips.2016.04.007 URL |
| [12] |
Rosati A, Leone A, Del Valle L, et al. Evidence for BAG3 modulation of HIV-1 gene transcription[J]. J Cell Physiol, 2007, 210(3):676-683.
doi: 10.1002/jcp.20865 URL |
| [13] |
Rosati A, Khalili K, Deshmane SL, et al. BAG3 protein regulates caspase-3 activation in HIV-1-infected human primary microglial cells[J]. J Cell Physiol, 2009, 218(2):264-267.
doi: 10.1002/jcp.21604 URL |
| [14] |
Kyratsous CA, Silverstein SJ. The co-chaperone BAG3 regulates Herpes Simplex Virus replication[J]. PNAS, 2008, 105(52):20912-20917.
doi: 10.1073/pnas.0810656105 pmid: 19088197 |
| [15] | 盛慧, 陈姗姗, 艾聪聪, 等. 利用酵母双杂交技术筛选卵菌效应因子互作蛋白概述[J]. 山东农业大学学报:自然科学版, 2019, 50(3):357-360. |
| Sheng H, Chen SS, Ai CC, et al. A review of interaction proteins of oomycete effectors screened by yeast two hybrid system[J]. J Shandong Agric Univ:Nat Sci Ed, 2019, 50(3):357-360. | |
| [16] |
Brooks D, Naeem F, Stetsiv M, et al. Drosophila NUAK functions with Starvin/BAG3 in autophagic protein turnover[J]. PLoS Genet, 2020, 16(4):e1008700.
doi: 10.1371/journal.pgen.1008700 URL |
| [17] | Meriin AB, Narayanan A, Meng L, et al. Hsp70-Bag3 complex is a hub for proteotoxicity-induced signaling that controls protein aggregation[J]. PNAS, 2018, 115(30):E7043-E7052. |
| [18] | Sherman MY, Gabai V. The role of Bag3 in cell signaling[J]. J Cell Biochem, 2021:jcb. 30111. |
| [19] |
Cirone M. ER stress, UPR activation and the inflammatory response to viral infection[J]. Viruses, 2021, 13(5):798.
doi: 10.3390/v13050798 URL |
| [20] |
Münz C. Autophagy proteins in viral exocytosis and anti-viral immune responses[J]. Viruses, 2017, 9(10):288.
doi: 10.3390/v9100288 URL |
| [21] | Mao JR, Lin E, He L, et al. Autophagy and viral infection[M]// Advances in Experimental Medicine and Biology. Singapore: Springer Singapore, 2019:55-78. |
| [22] |
Iyer K, Chand K, Mitra A, et al. Diversity in heat shock protein families:functional implications in virus infection with a comprehensive insight of their role in the HIV-1 life cycle[J]. Cell Stress Chaperones, 2021, 26(5):743-768.
doi: 10.1007/s12192-021-01223-3 URL |
| [23] |
Rao YL, Wan QY, Su H, et al. ROS-induced HSP70 promotes cytoplasmic translocation of high-mobility group box 1b and stimulates antiviral autophagy in grass carp kidney cells[J]. J Biol Chem, 2018, 293(45):17387-17401.
doi: 10.1074/jbc.RA118.003840 URL |
| [24] |
Klimek C, Jahnke R, Wördehoff J, et al. The Hippo network kinase STK38 contributes to protein homeostasis by inhibiting BAG3-mediated autophagy[J]. Biochim Biophys Acta Mol Cell Res, 2019, 1866(10):1556-1566.
doi: 10.1016/j.bbamcr.2019.07.007 URL |
| [25] |
Sharma A, Batra J, Stuchlik O, et al. Influenza A virus nucleoprotein activates the JNK stress-signaling pathway for viral replication by sequestering host filamin A protein[J]. Front Microbiol, 2020, 11:581867.
doi: 10.3389/fmicb.2020.581867 URL |
| [26] |
Dotson D, Woodruff EA, Villalta F, et al. Filamin A is involved in HIV-1 vpu-mediated evasion of host restriction by modulating tetherin expression[J]. J Biol Chem, 2016, 291(8):4236-4246.
doi: 10.1074/jbc.M115.708123 URL |
| [27] | Liu C, Qin XW, He J, et al. Roles of extracellular matrix components in Tiger frog virus attachment to fathead minnow(Pimephales promelas)cells[J]. Fish Shellfish Immunol, 2020, 107(pt a):9-15. |
| [28] |
Millen S, Gross C, Donhauser N, et al. Collagen IV(COL4A1, COL4A2), a component of the viral biofilm, is induced by the HTLV-1 oncoprotein tax and impacts virus transmission[J]. Front Microbiol, 2019, 10:2439.
doi: 10.3389/fmicb.2019.02439 URL |
| [29] |
Huang SZ, Liu K, Cheng AC, et al. SOCS proteins participate in the regulation of innate immune response caused by viruses[J]. Front Immunol, 2020, 11:558341.
doi: 10.3389/fimmu.2020.558341 URL |
| [30] |
Monie DD, Correia C, Zhang C, et al. Modular network mechanism of CCN1-associated resistance to HSV-1-derived oncolytic immunovirotherapies for glioblastomas[J]. Sci Rep, 2021, 11(1):11198.
doi: 10.1038/s41598-021-90718-1 URL |
| [31] |
Liu Y, Yin W, Wang JW, et al. KRAB-zinc finger protein ZNF268a deficiency attenuates the virus-induced pro-inflammatory response by preventing IKK complex assembly[J]. Cells, 2019, 8(12):1604.
doi: 10.3390/cells8121604 URL |
| [1] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [2] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [3] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [4] | 侯筱媛, 车郑郑, 李姮静, 杜崇玉, 胥倩, 王群青. 大豆膜系统cDNA文库的构建及大豆疫霉效应子PsAvr3a互作蛋白的筛选[J]. 生物技术通报, 2023, 39(4): 268-276. |
| [5] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [6] | 刘娟, 朱春晓, 肖雪琼, 莫陈汨, 王高峰, 肖炎农. 淡紫紫孢菌亲环蛋白PlCYP6 互作蛋白的筛选[J]. 生物技术通报, 2021, 37(7): 137-145. |
| [7] | 杨华杰, 周玉萍, 范甜, 吕天晓, 谢楚萍, 田长恩. 拟南芥IQM4互作蛋白的筛选和鉴定[J]. 生物技术通报, 2021, 37(11): 190-196. |
| [8] | 郝小花, 戴佳利, 暨文劲, 黄丹, 李东屏, 田连福. 水稻籽粒低镉蛋白LCD互作蛋白的筛选与鉴定[J]. 生物技术通报, 2020, 36(11): 21-29. |
| [9] | 钟李婷, 陈秀珍, 唐云, 李俊仁, 王小兵, 刘彦婷, 周璇璇, 詹若挺, 陈立凯. 广藿香FPPS重组蛋白表达及互作蛋白筛选分析[J]. 生物技术通报, 2019, 35(12): 10-15. |
| [10] | 贾建磊, 陈倩, 靳继鹏, 袁赞, 张利平. 绵羊BMPR1B基因真核表达及产物互作蛋白的鉴定[J]. 生物技术通报, 2019, 35(12): 94-104. |
| [11] | 卢晓颖, 黄宝松, 马骞, 陈刚, 王忠良, 黄建盛, ERICAMENYOGBE, 谢瑞涛, 邓文鑫. 杂交石斑鱼干扰素调节因子3(IRF3)基因的克隆及 表达分析[J]. 生物技术通报, 2019, 35(10): 144-151. |
| [12] | 王艺桥,赵新杰,牛芳芳,郭小华,杨博,江元清. 拟南芥中WRKY31转录因子的转录活性与互作蛋白分析[J]. 生物技术通报, 2018, 34(5): 101-109. |
| [13] | 李书鹏, 杨秀芬, 袁京京, 邱德文. 蛋白激发子Hrip1互作蛋白的筛选及其原核表达[J]. 生物技术通报, 2017, 33(6): 182-189. |
| [14] | 袁敏,王莉,葛伟娜,张岚. 拟南芥FD与14-3-3/GRF7蛋白的相互作用研究[J]. 生物技术通报, 2017, 33(5): 117-122. |
| [15] | 潘现飞,施泉,林新春,徐英武,曹友志. 利用酵母双杂交系统筛选雷竹SOC1相互作用蛋白[J]. 生物技术通报, 2016, 32(8): 207-213. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||