








生物技术通报 ›› 2023, Vol. 39 ›› Issue (6): 102-108.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1058
陈勇1, 李亚鑫2, 王亚瑄2, 梁露洁2, 冯思源2, 田国宝2,3( )
)
收稿日期:2022-08-25
出版日期:2023-06-26
发布日期:2023-07-07
通讯作者:
田国宝,博士,教授,研究方向:临床细菌耐药与感染;E-mail: tiangb@mail.sysu.edu.cn作者简介:陈勇,硕士,主管检验师,研究方向:临床细菌耐药与感染;E-mail: cy-yongchen@outlook.com
基金资助:
CHEN Yong1, LI Ya-xin2, WANG Ya-xuan2, LIANG Lu-jie2, FENG Si-yuan2, Tian Guo-bao2,3( )
)
Received:2022-08-25
Published:2023-06-26
Online:2023-07-07
摘要:
多黏菌素耐药基因mcr-1的出现为临床感染治疗带来了新的挑战。自其发现以来,已有6大洲61个不同的国家或地区报道了mcr-1的流行。为了遏制mcr-1的流行,我国农业农村部颁布了禁用多黏菌素作为饲料添加剂的禁令。尽管已有研究指出停用多黏菌素作为动物饲料添加剂可有效降低动物源、环境源和人源样本中mcr-1阳性菌的检出率,但是mcr-1在临床上仍呈低流行性的状态。截至目前已经发现34种mcr-1突变体和9种不同的MCR家族蛋白,未来是否会进化出流行率更高的MCR亚型也有待观察。关于mcr-1介导多黏菌素耐药的分子机制及其影响细菌细胞壁的机制也有新的成果不断出现。本文将对mcr-1的流行性、耐药机制及其对细菌适应性影响的分子机制3个方面的最新进展进行简要综述,以期为遏制多黏菌素耐药基因mcr-1的传播提供可参考依据。
陈勇, 李亚鑫, 王亚瑄, 梁露洁, 冯思源, 田国宝. MCR-1介导多黏菌素耐药性的分子机制研究进展[J]. 生物技术通报, 2023, 39(6): 102-108.
CHEN Yong, LI Ya-xin, WANG Ya-xuan, LIANG Lu-jie, FENG Si-yuan, Tian Guo-bao. Research Progress in the Molecular Mechanism of MCR-1 Mediated Polymyxin Resistance[J]. Biotechnology Bulletin, 2023, 39(6): 102-108.
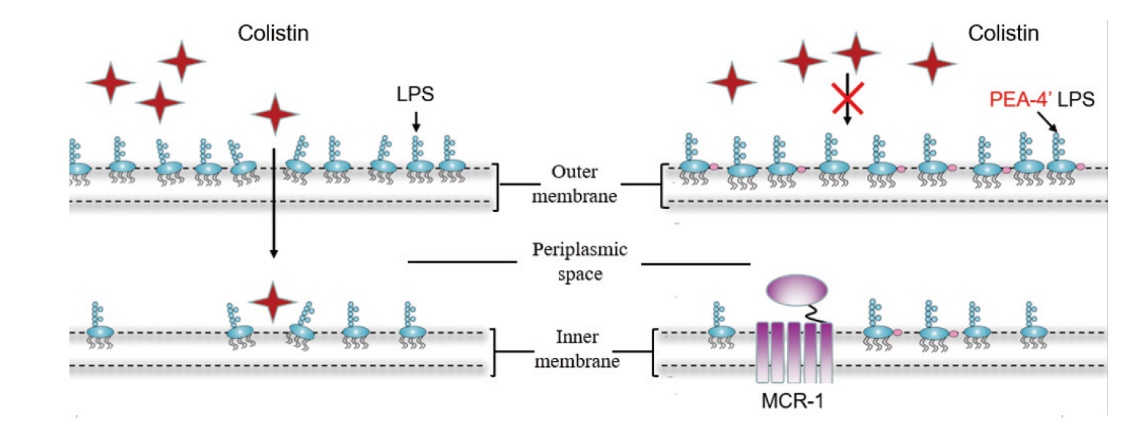
图1 MCR-1介导细菌产生多黏菌素的耐药机制 MCR-1将磷脂酰乙醇胺(PE)分子上带正电荷的磷酸乙醇胺(PEA)基团转移到细菌细胞外膜上的磷脂A,生成PEA-4' Lipid A,使细菌细胞膜上的正电荷增多,从而降低多黏菌素与脂多糖的亲和力,最终导致低水平耐药表型的产生
Fig. 1 Resistance mechanism of MCR-1-mediated bacterium producing polymyxin MCR-1 transfers the positively charged phosphoethanolamine(PEA)group on the phosphatidylethanolamine(PE)molecule to the phospholipid A on the bacterial cell outer membrane to generate PEA-4' Lipid A, which increases the positive charge on the bacterial cell membrane, thus reducing the affinity of polymyxin and lipopolysaccharide, and ultimately leads to the generation of low-level drug resistance phenotype
| [1] |
Talbot GH, Bradley J, Edwards JE Jr, et al. Bad bugs need drugs: an update on the development pipeline from the antimicrobial availability task force of the infectious diseases society of America[J]. Clin Infect Dis, 2006, 42(5): 657-668.
doi: 10.1086/499819 pmid: 16447111 |
| [2] |
Kaye KS, Pogue JM, Tran TB, et al. Agents of last resort: polymyxin resistance[J]. Infect Dis Clin North Am, 2016, 30(2): 391-414.
doi: 10.1016/j.idc.2016.02.005 URL |
| [3] |
Falagas ME, Kasiakou SK. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections[J]. Clin Infect Dis, 2005, 40(9): 1333-1341.
doi: 10.1086/429323 pmid: 15825037 |
| [4] |
Falagas ME, Rafailidis PI, Matthaiou DK. Resistance to polymyxins: mechanisms, frequency and treatment options[J]. Drug Resist Updat, 2010, 13(4/5): 132-138.
doi: 10.1016/j.drup.2010.05.002 URL |
| [5] |
Son SJ, Huang RJ, Squire CJ, et al. MCR-1: a promising target for structure-based design of inhibitors to tackle polymyxin resistance[J]. Drug Discov Today, 2019, 24(1): 206-216.
doi: S1359-6446(18)30025-4 pmid: 30036574 |
| [6] |
Li J, Nation RL, Milne RW, et al. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria[J]. Int J Antimicrob Agents, 2005, 25(1): 11-25.
doi: 10.1016/j.ijantimicag.2004.10.001 URL |
| [7] |
Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study[J]. Lancet Infect Dis, 2016, 16(2): 161-168.
doi: 10.1016/S1473-3099(15)00424-7 URL |
| [8] |
Shen ZQ, Wang Y, Shen YB, et al. Early emergence of mcr-1 in Escherichia coli from food-producing animals[J]. Lancet Infect Dis, 2016, 16(3): 293.
doi: 10.1016/S1473-3099(16)00061-X URL |
| [9] |
Haenni M, Poirel L, Kieffer N, et al. Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids[J]. Lancet Infect Dis, 2016, 16(3): 281-282.
doi: 10.1016/S1473-3099(16)00007-4 pmid: 26774244 |
| [10] |
Pham Thanh D, Thanh Tuyen H, Nguyen Thi Nguyen TT, et al. Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate[J]. J Antimicrob Chemother, 2016, 71(8): 2314-2317.
doi: 10.1093/jac/dkw173 pmid: 27246235 |
| [11] |
Jorgensen SB, Soraas A, Arnesen LS, et al. First environmental sample containing plasmid-mediated colistin-resistant ESBL-producing Escherichia coli detected in Norway[J]. APMIS, 2017, 125(9): 822-825.
doi: 10.1111/apm.2017.125.issue-9 URL |
| [12] |
Ling ZR, Yin WJ, Shen ZQ, et al. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9[J]. J Antimicrob Chemother, 2020, 75(11): 3087-3095.
doi: 10.1093/jac/dkaa205 pmid: 32514524 |
| [13] |
Nang SC, Li J, Velkov T. The rise and spread of mcr plasmid-mediated polymyxin resistance[J]. Crit Rev Microbiol, 2019, 45(2): 131-161.
doi: 10.1080/1040841X.2018.1492902 pmid: 31122100 |
| [14] |
Li RC, Xie MM, Zhang JF, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant[J]. J Antimicrob Chemother, 2017, 72(2): 393-401.
doi: 10.1093/jac/dkw411 pmid: 28073961 |
| [15] |
He YZ, Li XP, Miao YY, et al. The ISApl12 dimer circular intermediate participates in mcr-1 transposition[J]. Front Microbiol, 2019, 10: 15.
doi: 10.3389/fmicb.2019.00015 URL |
| [16] |
Sun J, Fang LX, Wu ZW, et al. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition[J]. Sci Rep, 2017, 7(1): 424.
doi: 10.1038/s41598-017-00095-x |
| [17] |
Li RC, Zhang P, Yang XR, et al. Identification of a novel hybrid plasmid coproducing MCR-1 and MCR-3 variant from an Escherichia coli strain[J]. J Antimicrob Chemother, 2019, 74(6): 1517-1520.
doi: 10.1093/jac/dkz058 URL |
| [18] |
Matamoros S, van Hattem JM, Arcilla MS, et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction[J]. Sci Rep, 2017, 7(1): 15364.
doi: 10.1038/s41598-017-15539-7 |
| [19] |
Wu RJ, Yi LX, Yu LF, et al. Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro[J]. Front Microbiol, 2018, 9: 331.
doi: 10.3389/fmicb.2018.00331 URL |
| [20] | Singh S, Pathak A, Kumar A, et al. Emergence of chromosome-borne colistin resistance gene mcr-1 in clinical isolates of Klebsiella pneumoniae from India[J]. Antimicrob Agents Chemother, 2018, 62(2): e01885-e01817. |
| [21] |
Lu XY, Xiao X, Liu Y, et al. Chromosome-mediated mcr-1 in Escherichia coli strain L73 from a goose[J]. Int J Antimicrob Agents, 2019, 54(1): 99-101.
doi: 10.1016/j.ijantimicag.2019.03.003 URL |
| [22] | Zhou HW, Zhang T, Ma JH, et al. Occurrence of plasmid- and chromosome-carried mcr-1 in waterborne Enterobacteriaceae in China[J]. Antimicrob Agents Chemother, 2017, 61(8): e00017-e00017. |
| [23] | Yamaguchi T, Kawahara R, Hamamoto K, et al. High prevalence of colistin-resistant Escherichia coli with chromosomally carried mcr-1 in healthy residents in Vietnam[J]. mSphere, 2020, 5(2): e00117-e00120. |
| [24] |
Wang Y, Xu CY, Zhang R, et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study[J]. Lancet Infect Dis, 2020, 20(10): 1161-1171.
doi: 10.1016/S1473-3099(20)30149-3 URL |
| [25] |
Liu YY, Zhou QL, He WY, et al. Mcr-1 and plasmid prevalence in Escherichia coli from livestock[J]. Lancet Infect Dis, 2020, 20(10): 1126.
doi: 10.1016/S1473-3099(20)30697-6 URL |
| [26] |
Shen C, Zhong LL, Yang YQ, et al. Dynamics of mcr-1 prevalence and mcr-1-positive Escherichia coli after the cessation of colistin use as a feed additive for animals in China: a prospective cross-sectional and whole genome sequencing-based molecular epidemiological study[J]. Lancet Microbe, 2020, 1(1): e34-e43.
doi: 10.1016/S2666-5247(20)30005-7 URL |
| [27] |
Shen C, Zhong LL, Zhong ZJ, et al. Prevalence of mcr-1 in colonized inpatients, China, 2011-2019[J]. Emerg Infect Dis, 2021, 27(9): 2502-2504.
doi: 10.3201/eid2709.203642 URL |
| [28] |
Usui M, Nozawa Y, Fukuda A, et al. Decreased colistin resistance and mcr-1 prevalence in pig-derived Escherichia coli in Japan after banning colistin as a feed additive[J]. J Glob Antimicrob Resist, 2021, 24: 383-386.
doi: 10.1016/j.jgar.2021.01.016 URL |
| [29] |
Fournier C, Aires-de-Sousa M, Nordmann P, et al. Occurrence of CTX-M-15- and MCR-1-producing Enterobacterales in pigs in Portugal: evidence of direct links with antibiotic selective pressure[J]. Int J Antimicrob Agents, 2020, 55(2): 105802.
doi: 10.1016/j.ijantimicag.2019.09.006 URL |
| [30] |
Sabnis A, Hagart KL, Klöckner A, et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane[J]. eLife, 2021, 10: e65836.
doi: 10.7554/eLife.65836 URL |
| [31] |
MacNair CR, Stokes JM, Carfrae LA, et al. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics[J]. Nat Commun, 2018, 9(1): 458.
doi: 10.1038/s41467-018-02875-z pmid: 29386620 |
| [32] |
Bialvaei AZ, Samadi Kafil H. Colistin, mechanisms and prevalence of resistance[J]. Curr Med Res Opin, 2015, 31(4): 707-721.
doi: 10.1185/03007995.2015.1018989 pmid: 25697677 |
| [33] |
Velkov T, Roberts KD, Nation RL, et al. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics[J]. Future Microbiol, 2013, 8(6): 711-724.
doi: 10.2217/fmb.13.39 pmid: 23701329 |
| [34] |
Chatzidimitriou M, Kavvada A, Kavvadas D, et al. Mcr genes conferring colistin resistance in enterobacterales; a five year overview[J]. Acta Med Acad, 2021, 50(3): 365-371.
doi: 10.5644/ama2006-124.355 pmid: 35164512 |
| [35] |
Hinchliffe P, Yang QE, Portal E, et al. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1[J]. Sci Rep, 2017, 7(1): 39392.
doi: 10.1038/srep39392 |
| [36] |
Coates K, Walsh TR, Spencer J, et al. 1.12 Å resolution crystal structure of the catalytic domain of the plasmid-mediated colistin resistance determinant MCR-2[J]. Acta Crystallogr Sect F, 2017, 73(8): 443-449.
doi: 10.1107/S2053230X17009669 URL |
| [37] | Sun J, Xu YC, Gao RS, et al. Deciphering MCR-2 colistin resistance[J]. mBio, 2017, 8(3): e00625-e00617. |
| [38] |
Xu YC, Lin JX, Cui T, et al. Mechanistic insights into transferable polymyxin resistance among gut bacteria[J]. J Biol Chem, 2018, 293(12): 4350-4365.
doi: 10.1074/jbc.RA117.000924 pmid: 29462787 |
| [39] |
Ma GX, Zhu YF, Yu ZC, et al. High resolution crystal structure of the catalytic domain of MCR-1[J]. Sci Rep, 2016, 6: 39540.
doi: 10.1038/srep39540 pmid: 28000749 |
| [40] |
Gao RS, Hu YF, Li ZC, et al. Dissemination and mechanism for the MCR-1 colistin resistance[J]. PLoS Pathog, 2016, 12(11): e1005957.
doi: 10.1371/journal.ppat.1005957 URL |
| [41] |
Zhang XF, Doi Y, Huang X, et al. Possible transmission of mcr-1-harboring Escherichia coli between companion animals and human[J]. Emerg Infect Dis, 2016, 22(9): 1679-1681.
doi: 10.3201/eid2209.160464 URL |
| [42] | Xu YC, Wei WH, Lei S, et al. An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance[J]. mBio, 2018, 9(2): e02317-e02317. |
| [43] |
Zhang HM, Srinivas S, Xu YC, et al. Genetic and biochemical mechanisms for bacterial lipid A modifiers associated with polymyxin resistance[J]. Trends Biochem Sci, 2019, 44(11): 973-988.
doi: S0968-0004(19)30135-5 pmid: 31279652 |
| [44] |
Sun J, Zhang HM, Liu YH, et al. Towards understanding MCR-like colistin resistance[J]. Trends Microbiol, 2018, 26(9): 794-808.
doi: S0966-842X(18)30042-8 pmid: 29525421 |
| [45] |
Xu YC, Chen HY, Zhang HM, et al. The MCR-3 inside linker appears as a facilitator of colistin resistance[J]. Cell Rep, 2021, 35(7): 109135.
doi: 10.1016/j.celrep.2021.109135 URL |
| [46] | Powers MJ, Trent MS. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics[J]. Proc Natl Acad Sci USA, 2018, 115(36): E8518-E8527. |
| [47] |
Wang XY, Quinn PJ, Yan AX. Kdo2-lipid A: structural diversity and impact on immunopharmacology[J]. Biol Rev, 2015, 90(2): 408-427.
doi: 10.1111/brv.2015.90.issue-2 URL |
| [48] |
Mandela E, Stubenrauch CJ, Ryoo D, et al. Adaptation of the periplasm to maintain spatial constraints essential for cell envelope processes and cell viability[J]. eLife, 2022, 11: e73516.
doi: 10.7554/eLife.73516 URL |
| [49] | Sutterlin HA, Shi HD, May KL, et al. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway[J]. Proc Natl Acad Sci USA, 2016, 113(11): E1565-E1574. |
| [50] |
Yang J, Wang HH, Lu YY, et al. A ProQ/FinO family protein involved in plasmid copy number control favours fitness of bacteria carrying mcr-1-bearing IncI2 plasmids[J]. Nucleic Acids Res, 2021, 49(7): 3981-3996.
doi: 10.1093/nar/gkab149 pmid: 33721023 |
| [51] |
Yang QE, Li M, Spiller OB, et al. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms[J]. Nat Commun, 2017, 8(1): 2054.
doi: 10.1038/s41467-017-02149-0 |
| [52] |
Yi LX, Durand R, Grenier F, et al. PixR, a novel activator of conjugative transfer of IncX4 resistance plasmids, mitigates the fitness cost of mcr-1 carriage in Escherichia coli[J]. mBio, 2022, 13(1): e0320921.
doi: 10.1128/mbio.03209-21 URL |
| [53] |
Feng SY, Liang WF, Li JC, et al. MCR-1-dependent lipid remodelling compromises the viability of Gram-negative bacteria[J]. Emerg Microbes Infect, 2022, 11(1): 1236-1249.
doi: 10.1080/22221751.2022.2065934 URL |
| [1] | 周振超, 郑吉, 帅馨怡, 林泽俊, 陈红. 高通量分析人类粪便、皮肤和水环境中共享抗生素抗性基因的分布[J]. 生物技术通报, 2023, 39(7): 288-297. |
| [2] | 董聪, 高庆华, 王玥, 罗同阳, 王庆庆. 基于联合策略提高FAD依赖的葡萄糖脱氢酶的酵母表达[J]. 生物技术通报, 2023, 39(6): 316-324. |
| [3] | 胡雪莹, 张越, 郭雅杰, 仇天雷, 高敏, 孙兴滨, 王旭明. 不同施肥处理农田土壤中噬菌体与细菌携带抗生素抗性基因的比较[J]. 生物技术通报, 2022, 38(9): 116-126. |
| [4] | 文畅, 刘晨, 卢诗韵, 许忠兵, 艾超凡, 廖汉鹏, 周顺桂. 一株新的多重耐药福氏志贺菌噬菌体生物学特性及基因组分析[J]. 生物技术通报, 2022, 38(9): 127-135. |
| [5] | 鲁兆祥, 王夕冉, 连新磊, 廖晓萍, 刘雅红, 孙坚. 基于功能宏基因组学挖掘抗生素耐药基因研究进展[J]. 生物技术通报, 2022, 38(9): 17-27. |
| [6] | 胡功政, 崔小蝶, 翟亚军, 贺丹丹. 细菌黏菌素耐药性及其逆转机制研究进展[J]. 生物技术通报, 2022, 38(9): 28-34. |
| [7] | 刘艺云, 邓利敏, 岳慧颖, 岳超, 刘健华. 质粒接合转移及其抑制剂的研究进展[J]. 生物技术通报, 2022, 38(9): 35-46. |
| [8] | 刘成程, 胡小芳, 冯友军. 细菌耐药:生化机制与应对策略[J]. 生物技术通报, 2022, 38(9): 4-16. |
| [9] | 刘理慧, 储锦华, 隋雨欣, 陈杨, 程古月. 沙门氏菌中主要毒力因子的研究进展[J]. 生物技术通报, 2022, 38(9): 72-83. |
| [10] | 李柳, 穆迎春, 刘璐, 张洪玉, 徐锦华, 杨臻, 乔璐, 宋金龙. 氟喹诺酮类抗生素及耐药基因污染控制的研究进展[J]. 生物技术通报, 2022, 38(9): 84-95. |
| [11] | 刘晓黎, 童真艺, 赵亮, 尹丽, 刘晨光. 非抗生素类活性物质抗幽门螺杆菌研究进展[J]. 生物技术通报, 2022, 38(9): 96-105. |
| [12] | 陈福暖, 黄瑜, 蔡佳, 王忠良, 简纪常, 王蓓. ABC转运蛋白结构及其在细菌致病性中的研究进展[J]. 生物技术通报, 2022, 38(6): 43-52. |
| [13] | 赵海晴, 李耘, 梁严内, 刘哲, 任亚林, 李金娟. 联合用药对嗜水气单胞菌耐药性影响研究进展[J]. 生物技术通报, 2022, 38(6): 53-65. |
| [14] | 朱浩, 张严伟, 刘润, 梁艳, 杨奕, 徐天乐, 杨章平. 抗生素佐剂与抗生素联用的抑菌作用研究进展[J]. 生物技术通报, 2022, 38(6): 66-73. |
| [15] | 许博楠, 冯佳, 周见庭, 蒋建兰. 无细胞蛋白合成系统中细胞提取物和模板DNA的研究进展[J]. 生物技术通报, 2022, 38(12): 100-114. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||