








生物技术通报 ›› 2023, Vol. 39 ›› Issue (8): 251-261.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1557
收稿日期:2022-12-27
出版日期:2023-08-26
发布日期:2023-09-05
通讯作者:
杨秀莲,女,博士,教授,研究方向:园林植物繁殖栽培与创新利用; E-mail: xly@njfu.edu.cn作者简介:付钰,男,硕士,研究方向:种质资源创新与利用; E-mail: 1091322721@qq.com
基金资助:
FU Yu( ), JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian(
), JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian( )
)
Received:2022-12-27
Published:2023-08-26
Online:2023-09-05
摘要:
嫁接是楸树(Catalpa bungei)良种扩繁的主要途径。为探讨以梓树和滇楸为砧木的两种‘苏楸1号’良种当年生嫁接苗生长差异,比较了二者的嫁接成活率、新梢长度、新梢粗度、接穗粗度/砧木粗度、叶面积等,以筛选较佳砧木,并通过高通量测序平台对两种嫁接苗叶片和顶芽进行转录组测序分析,采用实时荧光定量PCR技术对初步筛选的差异表达基因进行验证。结果显示,以梓树为砧木的嫁接成活率和嫁接苗各生长量均显著高于以滇楸为砧木的嫁接苗;转录组测序筛选出两个组合叶片和顶芽差异表达基因共计559个,其中上调表达基因192个,下调表达基因367个;GO富集分析表明559个差异表达基因显著富集在代谢过程、细胞过程、细胞器、结合和催化活性等功能类别;KEGG富集分析表明叶片中213个差异表达基因显著富集到66条代谢途径,顶芽中137个差异表达基因显著富集到45条代谢途径;初步筛选出的Unigene0040270、Unigene0006320、Unigene0036149和Unigene0007805等4个差异表达基因的RT-qPCR趋势与转录组数据一致。本研究结果发现两种嫁接苗的生长存在显著差异,其中以梓树为砧木的嫁接苗各生长参数更好,同时在转录组角度上对其差异进行了初步解析,为楸树嫁接分子研究提供一定的参考价值。
付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261.
FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome[J]. Biotechnology Bulletin, 2023, 39(8): 251-261.
| 嫁接组合 Grafting combination | 取样部位 Sampling part | 编号 Numbering |
|---|---|---|
| 苏楸1号/梓树 ‘Suqiu No. 1’/ Catalpa ovata | 叶片Leaf | ZS-L |
| 顶芽Bud | ZS-B | |
| 苏楸1号/滇楸 ‘Suqiu No. 1’/Catalpa fargesii | 叶片Leaf | DS-L |
| 顶芽Bud | DS-B |
表1 嫁接组合及取样部位
Table1 Grafting combination and sampling part
| 嫁接组合 Grafting combination | 取样部位 Sampling part | 编号 Numbering |
|---|---|---|
| 苏楸1号/梓树 ‘Suqiu No. 1’/ Catalpa ovata | 叶片Leaf | ZS-L |
| 顶芽Bud | ZS-B | |
| 苏楸1号/滇楸 ‘Suqiu No. 1’/Catalpa fargesii | 叶片Leaf | DS-L |
| 顶芽Bud | DS-B |
| 引物名称Primer name | 序列Sequence(5'-3') |
|---|---|
| CfMADH-F | AGCTTCCATTCTTTGCCTCA |
| CfMADH-R | TCCGAACAAAAGCAATACCC |
| Unigene0036149-F | AGGAACAGGAGAATGTGAAGAGG |
| Unigene0036149-R | CCGTCGTCACTTCTCGCAG |
| Unigene0007805-F | CAAAAAATGCTGGAAAAGTACCC |
| Unigene0007805-R | TGGTAAAATCGTGGTTCTCAAGTG |
| Unigene0040270-F | CCGCTTCATCTTGGTTCAGTG |
| Unigene0040270-R | AAGAAGGAAGATTTGCTGTTGGA |
| Unigene0006320-F | AGAAACACGAAAGTGAATGCTAAAC |
| Unigene0006320-R | GCACTCGGATGAAACGAAAAC |
表2 RT-qPCR引物序列
Table 2 Primer sequences of RT-qPCR
| 引物名称Primer name | 序列Sequence(5'-3') |
|---|---|
| CfMADH-F | AGCTTCCATTCTTTGCCTCA |
| CfMADH-R | TCCGAACAAAAGCAATACCC |
| Unigene0036149-F | AGGAACAGGAGAATGTGAAGAGG |
| Unigene0036149-R | CCGTCGTCACTTCTCGCAG |
| Unigene0007805-F | CAAAAAATGCTGGAAAAGTACCC |
| Unigene0007805-R | TGGTAAAATCGTGGTTCTCAAGTG |
| Unigene0040270-F | CCGCTTCATCTTGGTTCAGTG |
| Unigene0040270-R | AAGAAGGAAGATTTGCTGTTGGA |
| Unigene0006320-F | AGAAACACGAAAGTGAATGCTAAAC |
| Unigene0006320-R | GCACTCGGATGAAACGAAAAC |
| 嫁接组合 Grafting combination | 成活率 Survival rate/% | 新梢长度 New shoot length/cm | 新梢粗度 New shoot thickness/cm | 接穗粗度/砧木粗度 Scion diameter/Stock diameter | 叶面积 Leaf area/cm2 |
|---|---|---|---|---|---|
| ZS | 60.95±0.064** | 107.51±12.25** | 2.47±0.65** | 0.822±0.029** | 338.64±6.11** |
| DS | 32.05±0.038 | 83.43±5.06 | 2.04±1.57 | 0.721±0.087 | 247.61±6.59 |
表3 两个楸树嫁接苗的成活率和生长量
Table 3 Survival rates and growths of grafted seedlings of two Catalpa trees
| 嫁接组合 Grafting combination | 成活率 Survival rate/% | 新梢长度 New shoot length/cm | 新梢粗度 New shoot thickness/cm | 接穗粗度/砧木粗度 Scion diameter/Stock diameter | 叶面积 Leaf area/cm2 |
|---|---|---|---|---|---|
| ZS | 60.95±0.064** | 107.51±12.25** | 2.47±0.65** | 0.822±0.029** | 338.64±6.11** |
| DS | 32.05±0.038 | 83.43±5.06 | 2.04±1.57 | 0.721±0.087 | 247.61±6.59 |
| 高质量数据碱基总数 Clean data/bp | Q20/% | Q30/% | N/% | GC/% |
|---|---|---|---|---|
| 75 612 886 298 | 96.65-97.36 | 91.16-92.69 | 0 | 43.79-44.57 |
表4 12个样本测序数据的汇总结果
Table 4 Summary results of sequencing data for 12 samples
| 高质量数据碱基总数 Clean data/bp | Q20/% | Q30/% | N/% | GC/% |
|---|---|---|---|---|
| 75 612 886 298 | 96.65-97.36 | 91.16-92.69 | 0 | 43.79-44.57 |
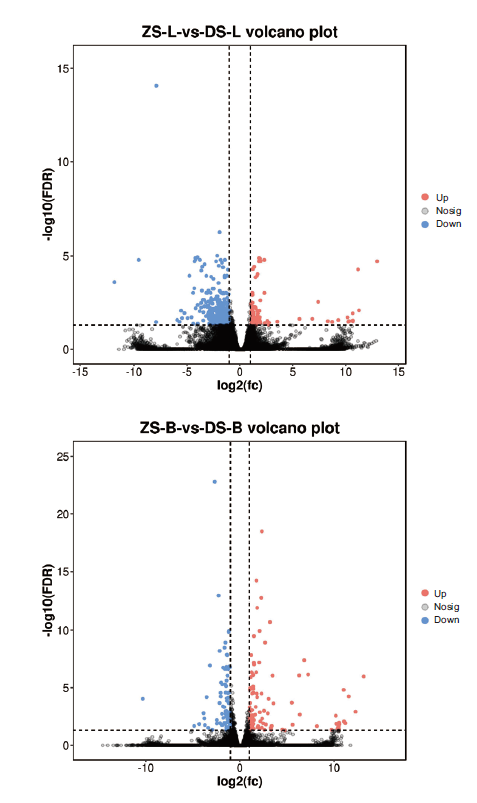
图2 不同嫁接组合差异基因火山图 横坐标表示两个分组间的差异倍数log2值,纵坐标表示两个分组差异的FDR的负Log10值,红色代表表达量上调,蓝色代表表达量下调
Fig. 2 Volcano plot of different genes in different grafting combinations The abscissa indicates the multiple log2 value of the difference between the two groups, the ordinate indicates the negative Log10 value of the FDR of the difference between the two groups. Red indicates the expression was up-regulated and blue was down-regulated
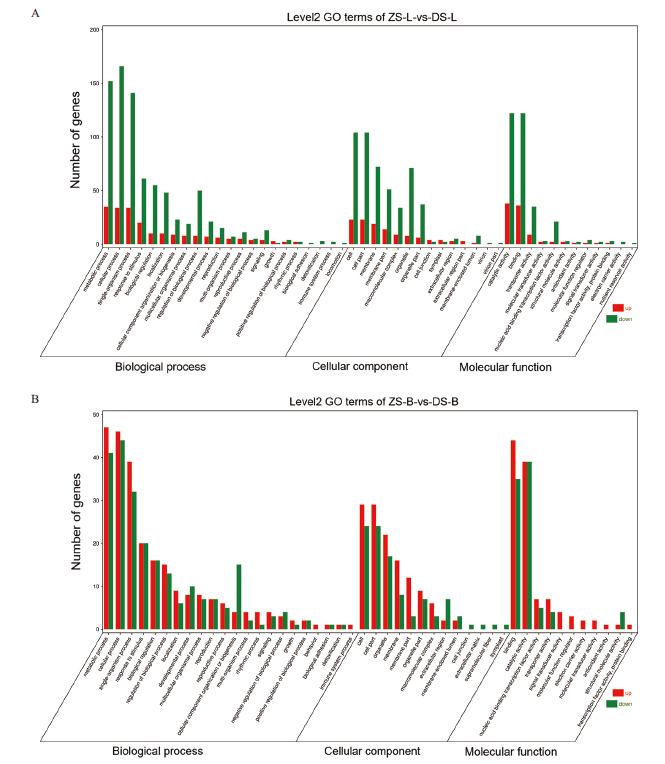
图3 不同嫁接组合差异基因的GO注释富集分析图 A:ZS-L vs DS-L;B:ZS-B vs DS-B。横坐标表示GO Term level 1与2,纵坐标表示差异表达基因的数量
Fig. 3 GO annotation enrichment analysis diagram of differential genes in different grafting combinations A:ZS-LvsDS-L; B:ZS-BvsDS-B. The abscissa represents GO Term level 1 and 2, the ordinate represents the number of differentially expressed genes
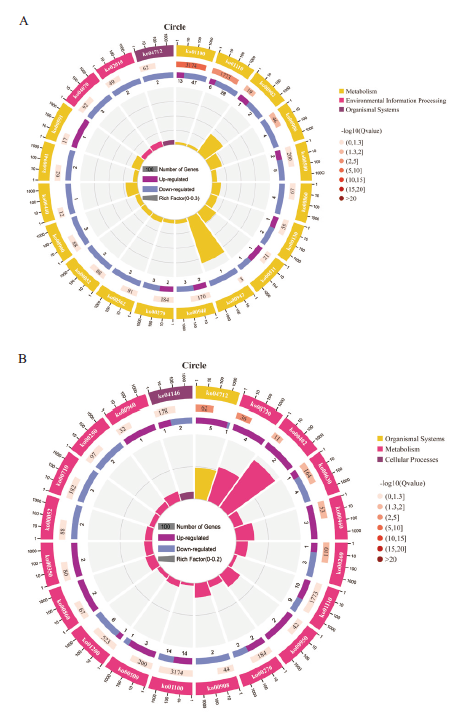
图4 不同嫁接组合显著差异基因KEGG富集分析图 A:ZS-L vs DS-L;B:ZS-B vs DS-B;第一圈:富集前20的pathway,圈外为差异基因数目的坐标尺。不同的颜色代表不同的A class;第二圈:差异基因背景中该pathway的数目以及Q值。差异基因背景数量越多条形越长,Q值越小颜色越红;第三圈:上下调差异基因比例条形图,深紫色代表上调差异基因比例,浅紫色代表下调差异基因比例;下方显示具体的数值;第四圈:各pathway的RichFactor值(该pathway中差异基因数量除以该pathway中所有数量),背景网格线,每一格代表0.1
Fig. 4 KEGG annotation enrichment analysis diagram of differential genes in different grafting combinations A:ZS-L vs DS-L;B:ZS-B vs DS-B. The first circle: enrich the top 20 pathways, outside the circle is the coordinate ruler of the number of differential genes. Different colors indicate different A classes. The second circle: the number and Q value of the pathway in the differential gene background. The more the number of differential genes background, the longer the bar, the smaller the Q value, the redder the color. The third circle: the bar graph of the ratio of up- and down-regulated differential genes, dark purple represents the ratio of up-regulated differential genes, and light purple represents the ratio of down-regulated differential genes; the lower part shows the specific value. The fourth circle: the RichFactor value of each pathway(the number of differential genes in the pathway divided by all the numbers in the pathway), background grid lines, each grid indicates 0.1
| 通路Pathway | 基因ID Unigene ID | 基因名称Symbol | 描述[ID]Description[ID] |
|---|---|---|---|
| 类胡萝卜素生物合成 Carotenoid biosynthesis | Unigene0051519 | PSY1 | 顺式八氢番茄红素合酶[EC:2.5.1.32]15-cis-Phytoene synthase |
| Unigene0046587 | LCY1 | 番茄红素β-环化酶[EC:5.5.1.19]Lycopene beta-cyclase | |
| Unigene0013377 | NCED2 | 9-顺式环氧类胡萝卜素双加氧酶[EC:1.13.11.51] 9-cis-Epoxycarotenoid dioxygenase | |
| Unigene0044139 | CYP707A2 | 脱落酸-8'-羟化酶[EC:1.14.14.137]Abscisic acid 8'-hydroxylase | |
| 卟啉和叶绿素代谢 Porphyrin and chlorophyll metabolism | Unigene0054531 | HEMA1 | 谷氨酰-tRNA还原酶[EC:1.2.1.70]Glutamyl-tRNA reductase |
| Unigene0017692 | CRD1 | 镁-原卟啉IX单甲酯(氧化)环化[EC:1.14.13.81] Magnesium-protoporphyrin IX monomethyl ester(oxidative)cyclase | |
| Unigene0005359 | CAO | 叶绿素a加氧酶[EC:1.14.13.122]Chlorophyllide a oxygenase | |
| Unigene0040270 | CAO | 叶绿素a加氧酶[EC:1.14.13.122]Chlorophyllide a oxygenase |
表5 与光合作用相关的差异表达基因统计
Tab.5 Statistics of differentially expressed genes related to photosynthesis
| 通路Pathway | 基因ID Unigene ID | 基因名称Symbol | 描述[ID]Description[ID] |
|---|---|---|---|
| 类胡萝卜素生物合成 Carotenoid biosynthesis | Unigene0051519 | PSY1 | 顺式八氢番茄红素合酶[EC:2.5.1.32]15-cis-Phytoene synthase |
| Unigene0046587 | LCY1 | 番茄红素β-环化酶[EC:5.5.1.19]Lycopene beta-cyclase | |
| Unigene0013377 | NCED2 | 9-顺式环氧类胡萝卜素双加氧酶[EC:1.13.11.51] 9-cis-Epoxycarotenoid dioxygenase | |
| Unigene0044139 | CYP707A2 | 脱落酸-8'-羟化酶[EC:1.14.14.137]Abscisic acid 8'-hydroxylase | |
| 卟啉和叶绿素代谢 Porphyrin and chlorophyll metabolism | Unigene0054531 | HEMA1 | 谷氨酰-tRNA还原酶[EC:1.2.1.70]Glutamyl-tRNA reductase |
| Unigene0017692 | CRD1 | 镁-原卟啉IX单甲酯(氧化)环化[EC:1.14.13.81] Magnesium-protoporphyrin IX monomethyl ester(oxidative)cyclase | |
| Unigene0005359 | CAO | 叶绿素a加氧酶[EC:1.14.13.122]Chlorophyllide a oxygenase | |
| Unigene0040270 | CAO | 叶绿素a加氧酶[EC:1.14.13.122]Chlorophyllide a oxygenase |
| 通路Pathway | 基因ID Unigene ID | 基因名称Symbol | 描述[ID]Description[ID] |
|---|---|---|---|
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | Unigene0038167 | SPS2 | 蔗糖-磷酸合酶[EC:2.4.1.14]Sucrose-phosphate synthase |
| Unigene0005917 | SS2 | 淀粉合酶[EC:2.4.1.21]Starch synthase | |
| Unigene0017971 | TPPJ | 海藻糖6-磷酸磷酸酶[EC:3.1.3.12]Trehalose 6-phosphate phosphatase | |
| Unigene0000390 | GLU3 | 内切葡聚糖酶[EC:3.2.1.4]Endoglucanase | |
| Unigene0022047 | GLU1 | β-葡萄糖苷酶[EC:3.2.1.21]beta-Glucosidase | |
| Unigene0048299 | INV*DC4 | β-果糖呋喃糖苷酶[EC:3.2.1.26]beta-Fructofuranosidase | |
| Unigene0006320 | WAXY | 颗粒结合淀粉合酶[EC:2.4.1.242]Granule-bound starch synthase | |
| Unigene0048994 | STP-1 | 糖原磷酸化酶[EC:2.4.1.1]Glycogen phosphorylase | |
| Unigene0047773 | BAM1 | β-淀粉酶[EC:3.2.1.2]beta-Amylase | |
| Unigene0043824 | BACOVA-02659 | β-葡萄糖苷酶[EC:3.2.1.21]beta-Glucosidase |
表6 与淀粉和蔗糖代谢相关的差异表达基因统计
Tab.6 Statistics of differentially expressed genes related to starch and sucrose metabolism
| 通路Pathway | 基因ID Unigene ID | 基因名称Symbol | 描述[ID]Description[ID] |
|---|---|---|---|
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | Unigene0038167 | SPS2 | 蔗糖-磷酸合酶[EC:2.4.1.14]Sucrose-phosphate synthase |
| Unigene0005917 | SS2 | 淀粉合酶[EC:2.4.1.21]Starch synthase | |
| Unigene0017971 | TPPJ | 海藻糖6-磷酸磷酸酶[EC:3.1.3.12]Trehalose 6-phosphate phosphatase | |
| Unigene0000390 | GLU3 | 内切葡聚糖酶[EC:3.2.1.4]Endoglucanase | |
| Unigene0022047 | GLU1 | β-葡萄糖苷酶[EC:3.2.1.21]beta-Glucosidase | |
| Unigene0048299 | INV*DC4 | β-果糖呋喃糖苷酶[EC:3.2.1.26]beta-Fructofuranosidase | |
| Unigene0006320 | WAXY | 颗粒结合淀粉合酶[EC:2.4.1.242]Granule-bound starch synthase | |
| Unigene0048994 | STP-1 | 糖原磷酸化酶[EC:2.4.1.1]Glycogen phosphorylase | |
| Unigene0047773 | BAM1 | β-淀粉酶[EC:3.2.1.2]beta-Amylase | |
| Unigene0043824 | BACOVA-02659 | β-葡萄糖苷酶[EC:3.2.1.21]beta-Glucosidase |
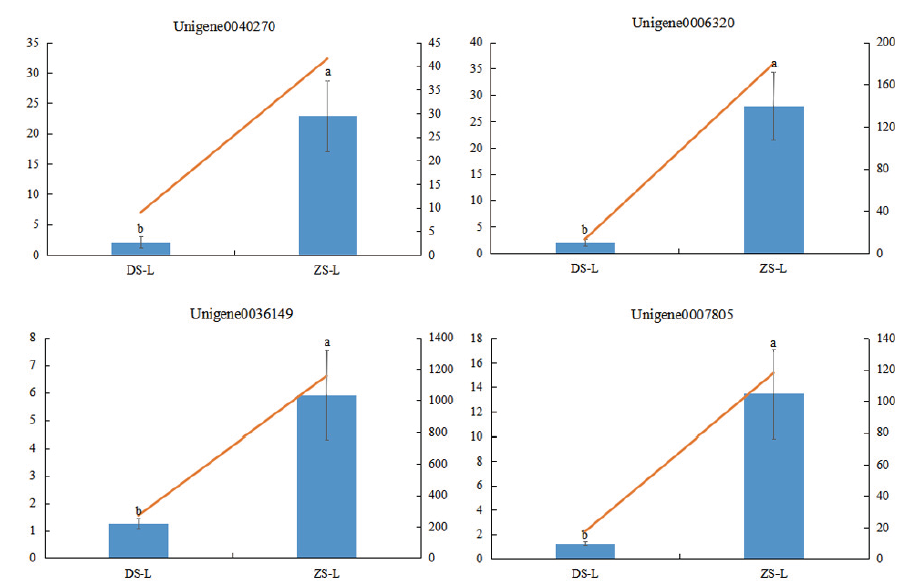
图7 四个基因在两个嫁接组合中的相对表达量 不同字母表示不同嫁接组合相对表达量差异显著(P<0.05)
Fig. 7 Relative expressions of four genes in two grafting combinations Different letters indicate the significant difference of relative expression at different grafting combinations.(P<0. 05)
| [1] |
Liu YM, Zhou L, Zhu YQ, et al. Anatomical features and its radial variations among different Catalpa bungei clones[J]. Forests, 2020, 11(8): 824.
doi: 10.3390/f11080824 URL |
| [2] |
Lu N, Zhang MM, Xiao Y, et al. Construction of a high-density genetic map and QTL mapping of leaf traits and plant growth in an interspecific F1 population of Catalpa bungei × Catalpa duclouxii Dode[J]. BMC Plant Biol, 2019, 19(1): 596.
doi: 10.1186/s12870-019-2207-y |
| [3] |
Wu JW, Li JY, Su Y, et al. A morphophysiological analysis of the effects of drought and shade on Catalpa bungei plantlets[J]. Acta Physiol Plant, 2017, 39(3): 80.
doi: 10.1007/s11738-017-2380-2 URL |
| [4] |
Xiao Y, Wang JH, Yun HL, et al. Genetic evaluation and combined selection for the simultaneous improvement of growth and wood properties in Catalpa bungei clones[J]. Forests, 2021, 12(7): 868.
doi: 10.3390/f12070868 URL |
| [5] | 王改萍, 王良桂, 王晓聪, 等. 楸树嫩枝扦插生根发育及根系特征分析[J]. 南京林业大学学报: 自然科学版, 2020, 44(6): 94-102. |
| Wang GP, Wang LG, Wang XC, et al. Dynamic characteristics of cutting rooting of Catalpa bungei with tender branches[J]. J Nanjing For Univ Nat Sci Ed, 2020, 44(6): 94-102. | |
| [6] |
Chen W, Meng PP, Feng H, et al. Effects of arbuscular mycorrhizal fungi on growth and physiological performance of Catalpa bungei C.A.Mey. under drought stress[J]. Forests, 2020, 11(10): 1117.
doi: 10.3390/f11101117 URL |
| [7] | Lin J, Wu LH, Jing L, et al. Effect of different plant growth regulators on callus induction in Catalpa bungei[J]. Afr J Agric Res, 2010, 5: 2699-2704. |
| [8] |
Miao L, Li Q, Sun TS, et al. Sugars promote graft union development in the heterograft of cucumber onto pumpkin[J]. Hortic Res, 2021, 8(1): 146.
doi: 10.1038/s41438-021-00580-5 |
| [9] |
Ren Y, Xu Q, Wang LW, et al. Involvement of metabolic, physiological and hormonal responses in the graft-compatible process of cucumber/pumpkin combinations was revealed through the integrative analysis of mRNA and miRNA expression[J]. Plant Physiol Biochem, 2018, 129: 368-380.
doi: 10.1016/j.plaphy.2018.06.021 URL |
| [10] | Karaca F, Yetİşİr H, Solmaz İ, et al. Rootstock potential of Turkish Lagenaria siceraria germplasm for watermelon: plant growth, yield and quality[J]. Turkish J Agric For, 2012: 167-177. |
| [11] |
Rasool A, Mansoor S, Bhat KM, et al. Mechanisms underlying graft union formation and rootstock scion interaction in horticultural plants[J]. Front Plant Sci, 2020, 11: 590847.
doi: 10.3389/fpls.2020.590847 URL |
| [12] |
Świerczyński S. The effect of rootstock activity for growth and root system soaking in Trichoderma atroviride on the graft success and continued growth of beech(Fagus sylvatica L.) plants[J]. Agronomy, 2022, 12(6): 1259.
doi: 10.3390/agronomy12061259 URL |
| [13] |
Liu WQ, Xiang CG, Li XJ, et al. Identification of long-distance transmissible mRNA between scion and rootstock in cucurbit seedling heterografts[J]. Int J Mol Sci, 2020, 21(15): 5253.
doi: 10.3390/ijms21155253 URL |
| [14] |
Kaseb MO, Umer MJ, Anees M, et al. Transcriptome profiling to dissect the role of genome duplication on graft compatibility mechanisms in watermelon[J]. Biology, 2022, 11(4): 575.
doi: 10.3390/biology11040575 URL |
| [15] |
Liu XY, Li J, Liu MM, et al. Transcriptome profiling to understand the effect of Citrus rootstocks on the growth of ‘shatangju’ mandarin[J]. PLoS One, 2017, 12(1): e0169897.
doi: 10.1371/journal.pone.0169897 URL |
| [16] |
Davoudi M, Song MF, Zhang MR, et al. Long-distance control of the scion by the rootstock under drought stress as revealed by transcriptome sequencing and mobile mRNA identification[J]. Hortic Res, 2022, 9: uhab033.
doi: 10.1093/hr/uhab033 URL |
| [17] | 申妍颖, 李雪涵, 李飞鸿, 等. 中间砧‘南通小方柿’嫁接柿转录组测序及分析[J]. 分子植物育种, 2019, 17(5): 1454-1466. |
| Shen YY, Li XH, Li FH, et al. Transcriptome sequencing and analysis of grafted persimmon with ‘Nantong-Xiaofangshi’ as the interstock[J]. Mol Plant Breed, 2019, 17(5): 1454-1466. | |
| [18] |
Li GF, Ma JJ, Tan M, et al. Transcriptome analysis reveals the effects of sugar metabolism and auxin and cytokinin signaling pathways on root growth and development of grafted apple[J]. BMC Genom, 2016, 17(1): 150.
doi: 10.1186/s12864-016-2484-x URL |
| [19] |
Young MD, Wakefield MJ, Smyth GK, et al. Gene ontology analysis for RNA-seq: accounting for selection bias[J]. Genome Biol, 2010, 11(2): R14.
doi: 10.1186/gb-2010-11-2-r14 URL |
| [20] |
Mao XZ, Cai T, Olyarchuk JG, et al. Automated genome annotation and pathway identification using the KEGG Orthology(KO)as a controlled vocabulary[J]. Bioinformatics, 2005, 21(19): 3787-3793.
doi: 10.1093/bioinformatics/bti430 URL |
| [21] | 杨英英, 赵林姣, 杨桂娟, 等. ‘麦缘锦楸’叶色表型RT-qPCR内参基因筛选及验证[J]. 林业科学研究, 2022, 35(1): 123-131. |
| Yang YY, Zhao LJ, Yang GJ, et al. Selection and validation of reference genes for leaf color phenotype in ‘maiyuanjinqiu’, a Catalpa fargesii variety, by RT-qPCR[J]. For Res, 2022, 35(1): 123-131. | |
| [22] |
He W, Xie R, Wang Y, et al. Comparative transcriptomic analysis on compatible/incompatible grafts in citrus[J]. Hortic Res, 2022, 9: uhab072.
doi: 10.1093/hr/uhab072 URL |
| [23] | 钟亚琴. 嫁接西瓜响应低钾胁迫的生理和分子机制研究[D]. 武汉: 华中农业大学, 2019. |
| Zhong YQ. Physiological and molecular mechanism of grafted watermelon in response to low potassium stress[D]. Wuhan: Huazhong Agricultural University, 2019. | |
| [24] | Thomas A, Brauer D, Sauer T, et al. Cultivar influences early rootstock and scion survival of grafted black walnut[J]. J Am Pomol Soc, 2008, 62(1): 3-12. |
| [25] |
Reig G, Zarrouk O, Font i Forcada C, et al. Anatomical graft compatibility study between apricot cultivars and different plum based rootstocks[J]. Sci Hortic, 2018, 237: 67-73.
doi: 10.1016/j.scienta.2018.03.035 URL |
| [26] | Huang Y, Zhao LQ, Kong QS, et al. Comprehensive mineral nutrition analysis of watermelon grafted onto two different rootstocks[J]. Hortic Plant J, 2016, 2(2): 105-113. |
| [27] | Bhuiyan MFA, Rahim MA, Alam MS. Study on the growth of plants produced by epicotyl(stone)grafting with different rootstock scion combinations in mango[J]. J Agrofor Environ, 2010, 3(2): 163-166. |
| [28] |
Harris ZN, Awale M, Bhakta N, et al. Multi-dimensional leaf phenotypes reflect root system genotype in grafted grapevine over the growing season[J]. GigaScience, 2021, 10(12): giab087.
doi: 10.1093/gigascience/giab087 URL |
| [29] | Yang YJ, Lu XM, Yan B, et al. Bottle gourd rootstock-grafting affects nitrogen metabolism in NaCl-stressed watermelon leaves and enhances short-term salt tolerance[J]. J Plant Physiol, 2013, 170(7): 653-661. |
| [30] |
Somkuwar RG, Bahetwar A, Khan I, et al. Changes in growth, photosynthetic activities, biochemical parameters and amino acid profile of Thompson Seedless grapes(Vitis vinifera L.)[J]. J Environ Biol, 2014, 35(6): 1157-1163.
pmid: 25522520 |
| [31] |
Proebsting WM, Maggard SP, Guo WW. The relationship of thiamine to the Alt locus of Pisum sativum L[J]. J Plant Physiol, 1990, 136(2): 231-235.
doi: 10.1016/S0176-1617(11)81671-8 URL |
| [32] |
Sayed SA, Gadallah MAA. Effects of shoot and root application of thiamin on salt-stressed sunflower plants[J]. Plant Growth Regul, 2002, 36(1): 71-80.
doi: 10.1023/A:1014784831387 URL |
| [33] |
Xu Q, Guo SR, Li L, et al. Proteomics analysis of compatibility and incompatibility in grafted cucumber seedlings[J]. Plant Physiol Biochem, 2016, 105: 21-28.
doi: 10.1016/j.plaphy.2016.04.001 URL |
| [34] |
Kim SH, Kim YH, Ahn YO, et al. Downregulation of the lycopene ϵ-cyclase gene increases carotenoid synthesis via the β-branch-specific pathway and enhances salt-stress tolerance in sweetpotato transgenic calli[J]. Physiol Plant, 2013, 147(4): 432-442.
doi: 10.1111/ppl.2013.147.issue-4 URL |
| [35] |
Komatsu A, Takanokura Y, Moriguchi T, et al. Differential expression of three sucrose-phosphate synthase isoforms during sucrose accumulation in citrus fruits(Citrus unshiu Marc.)[J]. Plant Science, 1999, 140(2): 169-178.
doi: 10.1016/S0168-9452(98)00217-9 URL |
| [36] |
Zhu YJ, Moore PH. Sucrose accumulation in the sugarcane stem is regulated by the difference between the activities of soluble acid invertase and sucrose phosphate synthase[J]. Plant Physiol, 1997, 115(2): 609-616.
doi: 10.1104/pp.115.2.609 pmid: 12223829 |
| [37] |
Pauly M, Keegstra K. Plant cell wall polymers as precursors for biofuels[J]. Curr Opin Plant Biol, 2010, 13(3): 304-311.
doi: 10.1016/j.pbi.2009.12.009 URL |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [3] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [4] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [5] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [6] | 杨洋, 朱金成, 娄慧, 韩泽刚, 张薇. 海岛棉与枯萎病菌的互作转录组分析[J]. 生物技术通报, 2023, 39(6): 259-273. |
| [7] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [8] | 谢洋, 邢雨蒙, 周国彦, 刘美妍, 银珊珊, 闫立英. 黄瓜二倍体及其同源四倍体果实转录组分析[J]. 生物技术通报, 2023, 39(3): 152-162. |
| [9] | 扈丽丽, 林柏荣, 王宏洪, 陈建松, 廖金铃, 卓侃. 最短尾短体线虫转录组及潜在效应蛋白分析[J]. 生物技术通报, 2023, 39(3): 254-266. |
| [10] | 孙言秋, 谢采芸, 汤岳琴. 耐高温酿酒酵母的构建与高温耐受机制解析[J]. 生物技术通报, 2023, 39(11): 226-237. |
| [11] | 徐俊, 叶雨晴, 牛雅静, 黄河, 张蒙蒙. 菊花根状茎发育的转录组分析[J]. 生物技术通报, 2023, 39(10): 231-245. |
| [12] | 罗皓天, 王龙, 王禹茜, 王月, 李佳祯, 杨梦珂, 张杰, 邓欣, 王红艳. 青狗尾草RNAi途径相关基因的全基因组鉴定和表达分析[J]. 生物技术通报, 2023, 39(1): 175-186. |
| [13] | 辛建攀, 李燕, 赵楚, 田如男. 镉胁迫下梭鱼草叶片转录组测序及苯丙烷代谢途径相关基因挖掘[J]. 生物技术通报, 2022, 38(6): 198-210. |
| [14] | 许瑾, 李涛, 李楚琳, 朱顺妮, 王忠铭, 向文洲. 温度对真眼点藻生长、总脂及二十碳五烯酸(EPA)合成的影响[J]. 生物技术通报, 2022, 38(6): 261-271. |
| [15] | 金姣姣, 刘自刚, 米文博, 徐明霞, 邹娅, 徐春梅, 赵彩霞. 利用RNA-Seq鉴定调控甘蓝型油菜叶片光合特性的低温胁迫应答基因[J]. 生物技术通报, 2022, 38(4): 126-142. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||