








生物技术通报 ›› 2024, Vol. 40 ›› Issue (2): 300-312.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0719
李亚男1( ), 张豪杰1, 梁梦静1, 罗涛1, 李旺宁1, 张春辉1, 季春丽1, 李润植1, 薛金爱1(
), 张豪杰1, 梁梦静1, 罗涛1, 李旺宁1, 张春辉1, 季春丽1, 李润植1, 薛金爱1( ), 崔红利1,2(
), 崔红利1,2( )
)
收稿日期:2023-07-23
出版日期:2024-02-26
发布日期:2024-03-13
通讯作者:
薛金爱,女,博士,教授,研究方向:生物技术与组学工程;E-mail: 306214803@qq.com;作者简介:李亚男,女,硕士研究生,研究方向:生物技术与组学工程;E-mail: Li_yanan_li@163.com
基金资助:
LI Ya-nan1( ), ZHANG Hao-jie1, LIANG Meng-jing1, LUO Tao1, LI Wang-ning1, ZHANG Chun-hui1, JI Chun-li1, LI Run-zhi1, XUE Jin-ai1(
), ZHANG Hao-jie1, LIANG Meng-jing1, LUO Tao1, LI Wang-ning1, ZHANG Chun-hui1, JI Chun-li1, LI Run-zhi1, XUE Jin-ai1( ), CUI Hong-li1,2(
), CUI Hong-li1,2( )
)
Received:2023-07-23
Published:2024-02-26
Online:2024-03-13
摘要:
【目的】钙依赖蛋白激酶(CDPK或CPK)广泛参与植物生长发育和环境胁迫响应。为探究CDPK基因在雨生红球藻(Haematococcus pluvialis)生长发育及虾青素积累中的功能,对HpCDPK基因家族进行鉴定和表达分析。【方法】利用生物信息学方法鉴定雨生红球藻HpCDPK基因家族成员,分析其编码蛋白的理化特性、系统进化、保守基序和功能结构域,以及缺氮等胁迫条件下HpCDPK成员基因的表达谱。【结果】结果表明,共鉴定7个HpCDPK基因家族成员,所有HpCDPK蛋白都含有典型EF-hand基序和蛋白激酶结构域。系统发育分析表明,这些HpCDPK与来自莱茵衣藻的CrCDPK聚为一类,与高等植物拟南芥的AtCDPK分开,暗示在进化过程中发生了种属特异性的基因复制事件。转录组数据分析表明,HpCDPK基因表达受到多种胁迫诱导,其中HpCDPK7基因转录水平在缺氮条件下上调最为明显。此外,HpCDPK与类胡萝卜素合成相关基因的表达相关性分析结果显示,HpCDPK与多个类胡萝卜素合成相关基因表达密切关联,特别是HpCDPK2与虾青素合成关键基因(BKT和BCH)的表达显著正相关(P<0.05)。【结论】本研究鉴定了7个HpCDPK基因,HpCDPK7基因在缺氮条件下的表达上调最为明显,HpCDPK2可能在类胡萝卜素及虾青素合成积累中发挥重要调控作用。研究结果为后续深入解析HpCDPK介导雨生红球藻胁迫响应和类胡萝卜素合成积累的功能及分子机制提供参考依据。
李亚男, 张豪杰, 梁梦静, 罗涛, 李旺宁, 张春辉, 季春丽, 李润植, 薛金爱, 崔红利. 雨生红球藻钙依赖蛋白激酶(CDPK)家族鉴定与表达分析[J]. 生物技术通报, 2024, 40(2): 300-312.
LI Ya-nan, ZHANG Hao-jie, LIANG Meng-jing, LUO Tao, LI Wang-ning, ZHANG Chun-hui, JI Chun-li, LI Run-zhi, XUE Jin-ai, CUI Hong-li. Identification and Expression Analysis of Calcium-dependent Protein Kinase(CDPK)Family in Haematococcus pluvialis[J]. Biotechnology Bulletin, 2024, 40(2): 300-312.
| 基因名称 Gene name | 基因编号 Gene No. | 蛋白长度 Protein length/aa | 分子量 MW/Da | 等电点 pI | EF手型数量 EF-hand number | N-肉豆蔻酰化 N- myristoylator | N-棕榈酰化 N-palmitoylation | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| HpCDPK1 | comp60148_c2 | 605 | 67 397.89 | 5.92 | 4 | No | Yes | Cytoplasmic |
| HpCDPK2 | comp60654_c0 | 545 | 59 849.50 | 6.08 | 4 | No | Yes | Chloroplast |
| HpCDPK3 | comp57813_c0 | 478 | 53 280.35 | 5.71 | 4 | No | No | Cytoplasmic |
| HpCDPK4 | comp60035_c0 | 676 | 73 764.11 | 6.03 | 2 | No | Yes | Cytoplasmic |
| HpCDPK5 | comp58900_c0 | 716 | 76 076.86 | 6.39 | 4 | No | Yes | Nuclear |
| HpCDPK6 | comp61638_c1 | 1 479 | 153 784.74 | 7.29 | 4 | No | Yes | Nuclear |
| HpCDPK7 | comp60433_c0 | 633 | 70 805.48 | 5.79 | 4 | Yes | Yes | Mitochondrial |
表1 雨生红球藻CDPK基因家族成员信息
Table 1 Members of CDPK gene family in H. pluvialis
| 基因名称 Gene name | 基因编号 Gene No. | 蛋白长度 Protein length/aa | 分子量 MW/Da | 等电点 pI | EF手型数量 EF-hand number | N-肉豆蔻酰化 N- myristoylator | N-棕榈酰化 N-palmitoylation | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| HpCDPK1 | comp60148_c2 | 605 | 67 397.89 | 5.92 | 4 | No | Yes | Cytoplasmic |
| HpCDPK2 | comp60654_c0 | 545 | 59 849.50 | 6.08 | 4 | No | Yes | Chloroplast |
| HpCDPK3 | comp57813_c0 | 478 | 53 280.35 | 5.71 | 4 | No | No | Cytoplasmic |
| HpCDPK4 | comp60035_c0 | 676 | 73 764.11 | 6.03 | 2 | No | Yes | Cytoplasmic |
| HpCDPK5 | comp58900_c0 | 716 | 76 076.86 | 6.39 | 4 | No | Yes | Nuclear |
| HpCDPK6 | comp61638_c1 | 1 479 | 153 784.74 | 7.29 | 4 | No | Yes | Nuclear |
| HpCDPK7 | comp60433_c0 | 633 | 70 805.48 | 5.79 | 4 | Yes | Yes | Mitochondrial |

图3 雨生红球藻HpCDPK家族成员的保守基序和功能结构域 不同的图案用特定的颜色表示;紫色框表示丝氨酸/苏氨酸蛋白激酶结构域(SM000220),绿色框表示EF-Hand(SM000054)
Fig. 3 Conserved motif and functional domain of HpCDPK family members of H. pluvialis Different motif is represent by specific color. Purple hollow box indictes the Serine/ Threonine protein kinases domain(SM000220), and green hollow box indicted the EF-hand(SM000054)

图4 MEME预测的雨生红球藻HpCDPK保守基序 横坐标是氨基酸的位置,纵坐标是氨基酸在一个部位的比例
Fig. 4 Conserved motifs of HpCDPKs in H. pluvialis predicted by MEME The abscissa indicates amino acid position and the ordinate indicates the proportion of amino acid at a site

图8 雨生红球藻HpCDPK基因在非生物胁迫下的表达模式 A:正常光(N),蓝光(B),高白光(W);B:正常光(L),正常光和紫外灯200 W(LU200), 正常光和紫外灯400 W(LU400), 黑暗(D),黑暗和紫外灯200 W(DU200),黑暗和紫外灯400 W(DU400);C:高光(CK),高光和α-酮戊二酸(OG),高光和缺氮(ND),高光、缺氮和α-酮戊二酸(ND-OG)
Fig. 8 Expression patterns of HpCDPK genes in H. pluvialis in response to abiotic stresses A: Normal light(N), blue light(B), and high white light(W). B: Normal light(L), normal light and UV-B 200 W(LU200), normal light and UV-B 400 W(LU400), dark(D), dark and UV-B 200 W(DU200), dark and UV-B 400 W(DU400). C: High light(CK), high light and α-ketoglutaric acid(OG), high light and nitrogen deficiency(ND), high light, nitrogen deficiency and α-ketoglutaric acid(ND-OG)
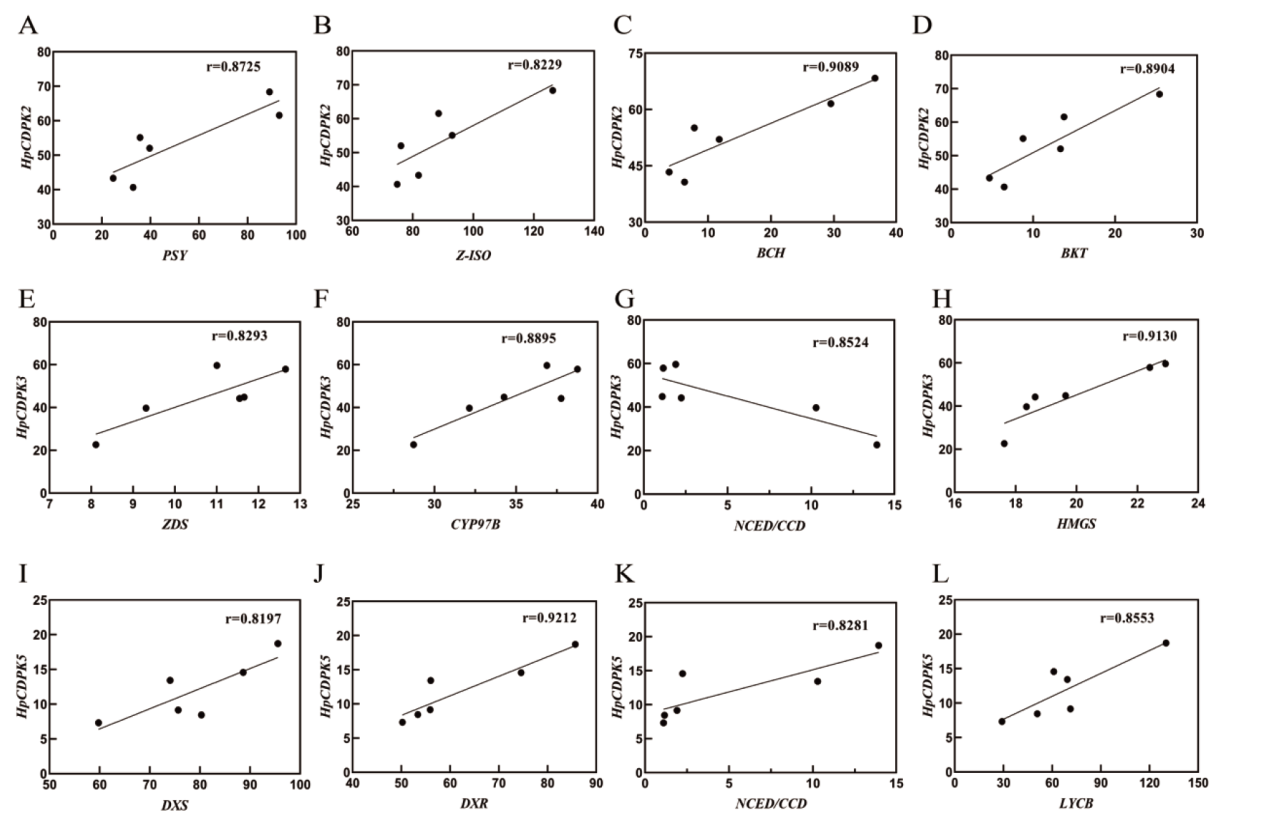
图10 HpCDPK2、HpCDPK3和HpCDPK5基因与类胡萝卜素及虾青素合成及积累基因的表达量相关性
Fig. 10 Correlation between HpCDPK2, HpCDPK3, HpCDPK5 genes and gene expressions for the biosynthesis and accumulation of carotenoid and astaxanthin
| [1] |
Ranty B, Aldon D, Cotelle V, et al. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses[J]. Front Plant Sci, 2016, 7: 327.
doi: 10.3389/fpls.2016.00327 pmid: 27014336 |
| [2] |
Hepler PK. Calcium: a central regulator of plant growth and development[J]. Plant Cell, 2005, 17(8): 2142-2155.
doi: 10.1105/tpc.105.032508 pmid: 16061961 |
| [3] |
Lecourieux D, Ranjeva R, Pugin A. Calcium in plant defence-signalling pathways[J]. New Phytol, 2006, 171(2): 249-269.
doi: 10.1111/j.1469-8137.2006.01777.x pmid: 16866934 |
| [4] |
Sanders D, Brownlee C, Harper JF. Communicating with calcium[J]. Plant Cell, 1999, 11(4): 691-706.
doi: 10.1105/tpc.11.4.691 URL |
| [5] |
Hamel LP, Sheen J, Séguin A. Ancient signals: comparative genomics of green plant CDPKs[J]. Trends Plant Sci, 2014, 19(2): 79-89.
doi: 10.1016/j.tplants.2013.10.009 URL |
| [6] | Cheng SH, Willmann MR, Chen HC, et al. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family[J]. Plant Physiol, 2002, 129(2): 469-485. |
| [7] |
Boudsocq M, Sheen J. CDPKs in immune and stress signaling[J]. Trends Plant Sci, 2013, 18(1): 30-40.
doi: 10.1016/j.tplants.2012.08.008 pmid: 22974587 |
| [8] |
Ludwig AA, Romeis T, Jones JDG. CDPK-mediated signalling pathways: specificity and cross-talk[J]. J Exp Bot, 2004, 55(395): 181-188.
doi: 10.1093/jxb/erh008 pmid: 14623901 |
| [9] |
Harmon AC, Gribskov M, Gubrium E, et al. The CDPK superfamily of protein kinases[J]. New Phytol, 2001, 151(1): 175-183.
doi: 10.1046/j.1469-8137.2001.00171.x pmid: 33873379 |
| [10] |
Yip Delormel T, Boudsocq M. Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana[J]. New Phytol, 2019, 224(2): 585-604.
doi: 10.1111/nph.16088 pmid: 31369160 |
| [11] |
Simeunovic A, Mair A, Wurzinger B, et al. Know where your clients are: subcellular localization and targets of calcium-dependent protein kinases[J]. J Exp Bot, 2016, 67(13): 3855-3872.
doi: 10.1093/jxb/erw157 pmid: 27117335 |
| [12] |
Hrabak EM, Chan CWM, Gribskov M, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases[J]. Plant Physiol, 2003, 132(2): 666-680.
doi: 10.1104/pp.102.011999 URL |
| [13] |
Li YJ, Fei XW, Dai HF, et al. Genome-wide identification of calcium-dependent protein kinases in Chlamydomonas reinhardtii and functional analyses in nitrogen deficiency-induced oil accumulation[J]. Front Plant Sci, 2019, 10: 1147.
doi: 10.3389/fpls.2019.01147 URL |
| [14] |
Romeis T, Ludwig AA, Martin R, et al. Calcium-dependent protein kinases play an essential role in a plant defence response[J]. EMBO J, 2001, 20(20): 5556-5567.
doi: 10.1093/emboj/20.20.5556 pmid: 11597999 |
| [15] |
Asano T, Tanaka N, Yang GX, et al. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice[J]. Plant Cell Physiol, 2005, 46(2): 356-366.
doi: 10.1093/pcp/pci035 URL |
| [16] |
Rutschmann F, Stalder U, Piotrowski M, et al. LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization[J]. Plant Physiol, 2002, 129(1): 156-168.
doi: 10.1104/pp.000869 pmid: 12011347 |
| [17] |
Kong XP, Lv W, Jiang SS, et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize[J]. BMC Genomics, 2013, 14: 433.
doi: 10.1186/1471-2164-14-433 pmid: 23815483 |
| [18] |
Huang K, Peng L, Liu YY, et al. Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response[J]. Biochem Biophys Res Commun, 2018, 498(1): 92-98.
doi: 10.1016/j.bbrc.2017.11.175 URL |
| [19] |
Urao T, Katagiri T, Mizoguchi T, et al. Two genes that encode Ca(2+)-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana[J]. Mol Gen Genet, 1994, 244(4): 331-340.
doi: 10.1007/BF00286684 URL |
| [20] |
Boudsocq M, Willmann MR, McCormack M, et al. Differential innate immune signalling via Ca2+ sensor protein kinases[J]. Nature, 2010, 464(7287): 418-422.
doi: 10.1038/nature08794 |
| [21] |
Zhang CH, Zhang LT, Liu JG. Exogenous sodium acetate enhances astaxanthin accumulation and photoprotection in Haematococcus pluvialis at the non-motile stage[J]. J Appl Phycol, 2019, 31(2): 1001-1008.
doi: 10.1007/s10811-018-1622-z |
| [22] |
Wang HM D, Li XC, Lee DJ, et al. Potential biomedical applications of marine algae[J]. Bioresour Technol, 2017, 244(Pt 2): 1407-1415.
doi: 10.1016/j.biortech.2017.05.198 URL |
| [23] |
Ren YY, Deng JQ, Huang JC, et al. Using green alga Haematococcus pluvialis for astaxanthin and lipid co-production: advances and outlook[J]. Bioresour Technol, 2021, 340: 125736.
doi: 10.1016/j.biortech.2021.125736 URL |
| [24] | 滕长英, 张立, 缪静, 等. 雨生红球藻虾青素积累机制的研究进展[J]. 海洋科学, 2006, 30(12): 77-81. |
| Teng CY, Zhang L, Miao J, et al. Review of astaxanthin accumulating mechanism in Haematococcus pluvialis[J]. Mar Sci, 2006, 30(12): 77-81. | |
| [25] |
Lamers J, van der Meer T, Testerink C. How plants sense and respond to stressful environments[J]. Plant Physiol, 2020, 182(4): 1624-1635.
doi: 10.1104/pp.19.01464 pmid: 32132112 |
| [26] | 崔红利, 许文鑫, 崔玉琳, 等. 光诱导雨生红球藻虾青素积累的信号通路转录组分析[J]. 生物工程学报, 2021, 37(4): 1260-1276. |
| Cui HL, Xu WX, Cui YL, et al. Transcriptome analysis of signal transduction pathway involved in light inducing astaxanthin accumulation in Haematococcus pluvialis[J]. Chin J Biotechnol, 2021, 37(4): 1260-1276. | |
| [27] | 许文鑫, 朱琴, 朱梅, 等. UV-B对雨生红球藻生长及虾青素积累的影响[J]. 水生生物学报, 2021, 45(6): 1281-1290. |
| Xu WX, Zhu Q, Zhu M, et al. Ultraviolet-b radiation enhances the growth and astaxanthin production in Haematococcus pluvialis[J]. Acta Hydrobiol Sin, 2021, 45(6): 1281-1290. | |
| [28] |
Johnson DR, Bhatnagar RS, Knoll LJ, et al. Genetic and biochemical studies of protein N-myristoylation[J]. Annu Rev Biochem, 1994, 63: 869-914.
pmid: 7979256 |
| [29] |
Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation[J]. J Biol Chem, 2001, 276(43): 39501-39504.
doi: 10.1074/jbc.R100042200 pmid: 11527981 |
| [30] |
Yuan M, Song ZH, Ying MD, et al. N-myristoylation: from cell biology to translational medicine[J]. Acta Pharmacol Sin, 2020, 41(8): 1005-1015.
doi: 10.1038/s41401-020-0388-4 pmid: 32203082 |
| [31] |
Zou JJ, Wei FJ, Wang C, et al. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress[J]. Plant Physiol, 2010, 154(3): 1232-1243.
doi: 10.1104/pp.110.157545 URL |
| [32] |
Li AL, Zhu YF, Tan XM, et al. Evolutionary and functional study of the CDPK gene family in wheat(Triticum aestivum L.)[J]. Plant Mol Biol, 2008, 66(4): 429-443.
doi: 10.1007/s11103-007-9281-5 URL |
| [33] |
Wang CT, Shao JM. Characterization of the ZmCK1 gene encoding a calcium-dependent protein kinase responsive to multiple abiotic stresses in maize[J]. Plant Mol Biol Rep, 2013, 31(1): 222-230.
doi: 10.1007/s11105-012-0496-5 URL |
| [34] |
Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains[J]. Science, 1988, 241(4861): 42-52.
doi: 10.1126/science.3291115 pmid: 3291115 |
| [35] |
Wen F, Ye F, Xiao ZL, et al. Genome-wide survey and expression analysis of calcium-dependent protein kinase(CDPK)in grass Brachypodium distachyon[J]. BMC Genomics, 2020, 21(1): 53.
doi: 10.1186/s12864-020-6475-6 |
| [36] | 任慧, 黄烯. 紫外光B波段光信号调控植物生长发育的研究进展[J]. 厦门大学学报: 自然科学版, 2021, 60(2): 327-338. |
| Ren H, Huang X. Research progress in the regulation of plant growth and development by ultraviolet-B light[J]. J Xiamen Univ Nat Sci, 2021, 60(2): 327-338. | |
| [37] |
Wargent JJ, Gegas VC, Jenkins GI, et al. UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation[J]. New Phytol, 2009, 183(2): 315-326.
doi: 10.1111/j.1469-8137.2009.02855.x pmid: 19402876 |
| [38] |
Fasano R, Gonzalez N, Tosco A, et al. Role of Arabidopsis UV RESISTANCE LOCUS 8 in plant growth reduction under osmotic stress and low levels of UV-B[J]. Mol Plant, 2014, 7(5): 773-791.
doi: 10.1093/mp/ssu002 URL |
| [39] |
Liang T, Yang Y, Liu HT. Signal transduction mediated by the plant UV-B photoreceptor UVR8[J]. New Phytol, 2019, 221(3): 1247-1252.
doi: 10.1111/nph.15469 pmid: 30315741 |
| [40] |
Ramani S, Chelliah J. UV-B-induced signaling events leading to enhanced-production of catharanthine in Catharanthus roseus cell suspension cultures[J]. BMC Plant Biol, 2007, 7: 61.
doi: 10.1186/1471-2229-7-61 URL |
| [41] |
Frattini M, Morello L, Breviario D. Rice calcium-dependent protein kinase isoforms OsCDPK2 and OsCDPK11 show different responses to light and different expression patterns during seed development[J]. Plant Mol Biol, 1999, 41(6): 753-764.
pmid: 10737140 |
| [42] |
Disch A, Schwender J, Müller C, et al. Distribution of the mevalonate and glyceraldehyde phosphate/pyruvate pathways for isoprenoid biosynthesis in unicellular algae and the cyanobacterium Synechocystis PCC 6714[J]. Biochem J, 1998, 333(Pt 2)(Pt 2):381-388.
doi: 10.1042/bj3330381 URL |
| [43] | Gilroy S, Trewavas A. Signal processing and transduction in plant cells: the end of the beginning?[J]. Nat Rev Mol Cell Biol, 2001, 2(4): 307-314. |
| [44] |
Pecker I, Gabbay R, Cunningham FX Jr, et al. Cloning and characterization of the cDNA for lycopene beta-cyclase from tomato reveals decrease in its expression during fruit ripening[J]. Plant Mol Biol, 1996, 30(4): 807-819.
doi: 10.1007/BF00019013 pmid: 8624411 |
| [45] |
Cunningham FX Jr, Gantt E. One ring or two? Determination of ring number in carotenoids by lycopene epsilon-cyclases[J]. Proc Natl Acad Sci USA, 2001, 98(5): 2905-2910.
doi: 10.1073/pnas.051618398 pmid: 11226339 |
| [46] | 刘贯山, 陈珈. 钙依赖蛋白激酶(CDPKs)在植物钙信号转导中的作用[J]. 植物学通报, 2003, 38(2): 160-167. |
| Liu GS, Chen J. Roles of calcium dependent protein kinases(CDPKs)in plant calcium signal transduction[J]. Chin Bull Bot, 2003, 38(2): 160-167. |
| [1] | 吴翠翠, 肖水平. 陆地棉HD-Zip家族全基因组鉴定及响应非生物胁迫的表达分析[J]. 生物技术通报, 2024, 40(2): 130-145. |
| [2] | 饶紫环, 谢志雄. 一株Olivibacter jilunii 纤维素降解菌株的分离鉴定与降解能力分析[J]. 生物技术通报, 2023, 39(8): 283-290. |
| [3] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [4] | 潘国强, 吴思源, 刘璐, 郭惠明, 程红梅, 苏晓峰. 大丽轮枝菌(Verticillim dahliae)突变体库的构建与分析[J]. 生物技术通报, 2023, 39(5): 112-119. |
| [5] | 任丽, 乔舒婷, 葛晨辉, 魏梓桐, 徐晨曦. 菠菜PSY基因家族的鉴定与表达分析[J]. 生物技术通报, 2023, 39(12): 169-178. |
| [6] | 陈广霞, 李秀杰, 蒋锡龙, 单雷, 张志昌, 李勃. 植物小分子信号肽参与非生物逆境胁迫应答的研究进展[J]. 生物技术通报, 2023, 39(11): 61-73. |
| [7] | 韩芳英, 胡昕, 王楠楠, 谢裕红, 王晓艳, 朱强. DREBs响应植物非生物逆境胁迫研究进展[J]. 生物技术通报, 2023, 39(11): 86-98. |
| [8] | 尤垂淮, 谢津津, 张婷, 崔天真, 孙欣路, 臧守建, 武奕凝, 孙梦瑶, 阙友雄, 苏亚春. 钩吻脂氧合酶基因 GeLOX1 的鉴定及低温胁迫表达分析[J]. 生物技术通报, 2023, 39(11): 318-327. |
| [9] | 杨佳宝, 周至铭, 张展, 冯丽, 孙黎. 向日葵HaLACS1的克隆、表达及酵母功能互补鉴定[J]. 生物技术通报, 2022, 38(6): 147-156. |
| [10] | 林科运, 段钰晶, 王高升, 孙念礼, 方玉洁, 王幼平. 甘蓝型油菜BnNF-YA1的克隆和功能鉴定[J]. 生物技术通报, 2022, 38(4): 106-116. |
| [11] | 张业猛, 朱丽丽, 陈志国. 藜麦NHX基因家族鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2022, 38(12): 184-193. |
| [12] | 曹映辉, 胡美娟, 童妍, 张燕萍, 赵凯, 彭东辉, 周育真. 建兰ABC基因家族鉴定及其在花发育过程中的表达模式分析[J]. 生物技术通报, 2022, 38(11): 162-174. |
| [13] | 魏英, 罗萌, 戴良英, 彭德良, 刘敬. 植物寄生线虫钙网蛋白的研究进展[J]. 生物技术通报, 2021, 37(7): 81-87. |
| [14] | 郝向阳, 刘范, 武欢, 王斌, 孙雪丽, 项蕾蕾, 王天池, 赖钟雄, 程春振. 非洲菊GjPAL的克隆及表达分析[J]. 生物技术通报, 2021, 37(6): 13-23. |
| [15] | 林艳丽, 覃建兵, 伍翔, 王岩岩, 潘佑找, 柳忠玉. 虎杖PcMYB1启动子的克隆及其活性分析[J]. 生物技术通报, 2021, 37(5): 48-55. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||