








生物技术通报 ›› 2024, Vol. 40 ›› Issue (2): 80-89.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0758
郑巧1,2( ), 林华1, 徐浩3, 安微1, 薛昌华1, 张婧1(
), 林华1, 徐浩3, 安微1, 薛昌华1, 张婧1( ), 韩国全2(
), 韩国全2( )
)
收稿日期:2023-08-09
出版日期:2024-02-26
发布日期:2024-03-13
通讯作者:
张婧,女,博士,高级农艺师,研究方向:分子生物学;E-mail: zjeane@163.com;作者简介:郑巧,女,硕士研究生,研究方向:食品质量安全检测与控制;E-mail: zq950222@163.com
基金资助:
ZHENG Qiao1,2( ), LIN Hua1, XU Hao3, AN Wei1, XUE Chang-hua1, ZHANG Jing1(
), LIN Hua1, XU Hao3, AN Wei1, XUE Chang-hua1, ZHANG Jing1( ), HAN Guo-quan2(
), HAN Guo-quan2( )
)
Received:2023-08-09
Published:2024-02-26
Online:2024-03-13
摘要:
【目的】为建立精准、高效的多重微滴式数字 PCR(droplet digital PCR,ddPCR)定量分析检测方法,开发可同时鉴别新型冠状病毒(severe acute respiratory syndrome coronavirus 2,SARS-CoV-2)ORF1ab 基因、N 基因、E 基因以及 Omicron 变异株 S 基因的检测体系,提高病毒性传染疾病的诊断效率及传播风险监测能力。【方法】通过筛选保守基因序列并设计特异性引物与探针,优化反应体系和扩增程序,对该方法的特异性、灵敏度及稳定性进行评价。以临床样本为实验材料,利用建立的 ddPCR 方法进行检测和验证,确定阳性检出率。【结果】多重 ddPCR 反应体系中,各对引物探针对目的片段均能有效扩增,SARS-CoV-2 ORF1ab 基因、N 基因、E 基因以及 Omicron 变异株 S 基因灵敏度检测下限分别为0.59、0.68、1.44、1.03 copies/µL。在20份临床样本的核酸检测中,共检出16份阳性样本,阳性率达80%(16/20),经荧光定量 PCR 方法复测符合率一致。【结论】研究建立的多重 ddPCR 方法特异性强、灵敏度高,可实现对临床样本中微量新冠病毒的精确定量检测。
郑巧, 林华, 徐浩, 安微, 薛昌华, 张婧, 韩国全. SARS-CoV-2 Omicron 变异株多重微滴式数字 PCR 定量方法的建立及应用[J]. 生物技术通报, 2024, 40(2): 80-89.
ZHENG Qiao, LIN Hua, XU Hao, AN Wei, XUE Chang-hua, ZHANG Jing, HAN Guo-quan. Establishment and Application of Multiplex Droplet Digital PCR for SARS-CoV-2 Omicron Variant[J]. Biotechnology Bulletin, 2024, 40(2): 80-89.
| 引物与探针Primer and probe | 序列Sequence(5'-3') | 产物长度Product length/bp |
|---|---|---|
| ORF1ab-F | TGTGCTAATGACCCTGT | 138 |
| ORF1ab-R | GTTTAAAAACGATTGTGCATCAG | |
| ORF1ab-P | FAM-AACTCCGCGAACCCATGCTT-BHQ1 | |
| N-F | GTCAAGCCTCTTCTCGTT | 101 |
| N-R | ATTGCCAGCCATTCTAGCAG | |
| N-P | HEX-CCTCATCACGTAGTCGCAACAGT-BHQ1 | |
| E-F | TTTCTTGCTTTCGTGGT | 132 |
| E-R | TTTAACACGAGAGTAAACGTA | |
| E-P | ROX-CTTCGATTGTGTGCGTACTGCT-BHQ2 | |
| S-F | GTTGCTGATTATTCTGTCCTA | 103 |
| S-R | AGACATTAGTAAAGCAGAGAT | |
| S-P | CY5-ACACTTAAAAGCGAAAAATGGT-MGB |
表1 引物与探针序列
Table 1 Primer and probe sequences
| 引物与探针Primer and probe | 序列Sequence(5'-3') | 产物长度Product length/bp |
|---|---|---|
| ORF1ab-F | TGTGCTAATGACCCTGT | 138 |
| ORF1ab-R | GTTTAAAAACGATTGTGCATCAG | |
| ORF1ab-P | FAM-AACTCCGCGAACCCATGCTT-BHQ1 | |
| N-F | GTCAAGCCTCTTCTCGTT | 101 |
| N-R | ATTGCCAGCCATTCTAGCAG | |
| N-P | HEX-CCTCATCACGTAGTCGCAACAGT-BHQ1 | |
| E-F | TTTCTTGCTTTCGTGGT | 132 |
| E-R | TTTAACACGAGAGTAAACGTA | |
| E-P | ROX-CTTCGATTGTGTGCGTACTGCT-BHQ2 | |
| S-F | GTTGCTGATTATTCTGTCCTA | 103 |
| S-R | AGACATTAGTAAAGCAGAGAT | |
| S-P | CY5-ACACTTAAAAGCGAAAAATGGT-MGB |

图1 SARS-CoV-2 ORF1ab 基因、N 基因、E 基因、S 基因扩增产物电泳图 M:DL2000;1:ORF1ab 基因扩增产物;2:N 基因扩增产物;3:E 基因扩增产物;4:S 基因扩增产物
Fig. 1 Electrophoresis map of SARS-CoV-2 ORF1ab, N, E, and S gene amplification products M: DL2000; 1: ORF1ab gene amplified product; 2: N gene amplified products; 3: E gene amplified products; 4: S gene amplified product

图2 SARS-CoV-2 ORF1ab 基因、N 基因、E 基因、S 基因单重ddPCR 检测一维图 A:ORF1ab 阳性质粒标准品,蓝色液滴;B:N 基因阳性质粒标准品,红色液滴;C:E 基因阳性质粒标准品,绿色液滴;D:S 基因阳性质粒标准品,黄色液滴;空白对照,灰色液滴
Fig. 2 One-dimensional map of SARS-CoV-2 ORF1ab gene, N gene, E gene and S gene by single ddPCR A: ORF1ab positive plasmid standard, blue droplets; B: N gene positive plasmid standard, red droplet; C: E gene positive plasmid standard, green droplet; D: S gene positive plasmid standard, yellow droplet; blank control, grey droplets

图3 四重ddPCR 特异性检测一维图 1F:Beta株;1G:Gamma株;1H:Delta株;2A:SARS-CoV-2 阳性质粒标准品;2B:猪流行性腹泻病毒;2C:猪传染性胃肠炎病毒;2D:甲型流感病毒;2E:诺如病毒;2F:腺病毒;2G:轮状病毒;2H:空白对照
Fig. 3 One-dimensional map of specificity detected by quadruple ddPCR 1F: Beta strain; 1G: Gamma strain; 1H: Delta strain; 2A: SARS-CoV-2 positive plasmid standard; 2B: porcine epidemic diarrhea virus; 2C: transmissible gastroenteritis of swine virus; 2D: influenza A virus; 2E: norovirus; 2F: adenovirus; 2G: rotavirus; 2H: blank control
| 基因名称 Gene name | 稀释度 Dilution | ddPCR 检测结果Results of ddPCR/(Cp·μL-1) | 平均值AVG | 标准差SD | 相对标准偏差RSD/% | ||
|---|---|---|---|---|---|---|---|
| Repeat 1 | Repeat 2 | Repeat 3 | |||||
| ORF1ab | 10-6 | 11230.63 | 11200.05 | 11315.73 | 11248.80 | 48.94 | 0.44 |
| 10-7 | 1163.21 | 1149.84 | 1172.26 | 1161.77 | 9.21 | 0.79 | |
| 10-8 | 96.43 | 88.66 | 89.98 | 91.69 | 3.39 | 3.70 | |
| 10-9 | 8.83 | 6.57 | 7.33 | 7.58 | 0.94 | 12.39 | |
| 10-10 | 0.74 | 0.44 | 0.58 | 0.59 | 0.12 | 20.89 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
| N | 10-6 | 6133.75 | 6226.00 | 6081.97 | 6147.21 | 59.57 | 0.97 |
| 10-7 | 617.10 | 621.28 | 598.72 | 612.37 | 9.80 | 1.60 | |
| 10-8 | 51.33 | 57.58 | 52.90 | 53.94 | 2.62 | 4.92 | |
| 10-9 | 5.33 | 5.69 | 4.40 | 5.14 | 0.54 | 10.57 | |
| 10-10 | 0.82 | 0.78 | 0.44 | 0.68 | 0.17 | 25.07 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
| E | 10-6 | 4350.43 | 4378.88 | 4334.00 | 4387.77 | 47.95 | 1.09 |
| 10-7 | 461.63 | 443.83 | 441.59 | 449.02 | 8.97 | 2.00 | |
| 10-8 | 45.01 | 50.60 | 43.13 | 46.25 | 3.17 | 6.86 | |
| 10-9 | 4.38 | 4.87 | 4.03 | 4.43 | 0.34 | 7.78 | |
| 10-10 | 1.32 | 1.69 | 1.32 | 1.44 | 0.17 | 12.08 | |
| 10-11 | 0 | 0.44 | 0 | 0.15 | 0.21 | 141.42 | |
| S | 10-6 | 5891.09 | 5839.83 | 5942.93 | 5891.28 | 42.09 | 0.71 |
| 10-7 | 591.21 | 558.87 | 593.71 | 581.26 | 15.87 | 2.73 | |
| 10-8 | 56.59 | 55.57 | 60.52 | 57.56 | 2.13 | 3.71 | |
| 10-9 | 5.06 | 5.87 | 5.92 | 5.62 | 0.39 | 7.02 | |
| 10-10 | 0.88 | 0.88 | 1.32 | 1.03 | 0.21 | 20.20 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
表2 四重 ddPCR 灵敏度与稳定性检测结果
Table 2 Sensitivity and stability detection results by quadruple ddPCR
| 基因名称 Gene name | 稀释度 Dilution | ddPCR 检测结果Results of ddPCR/(Cp·μL-1) | 平均值AVG | 标准差SD | 相对标准偏差RSD/% | ||
|---|---|---|---|---|---|---|---|
| Repeat 1 | Repeat 2 | Repeat 3 | |||||
| ORF1ab | 10-6 | 11230.63 | 11200.05 | 11315.73 | 11248.80 | 48.94 | 0.44 |
| 10-7 | 1163.21 | 1149.84 | 1172.26 | 1161.77 | 9.21 | 0.79 | |
| 10-8 | 96.43 | 88.66 | 89.98 | 91.69 | 3.39 | 3.70 | |
| 10-9 | 8.83 | 6.57 | 7.33 | 7.58 | 0.94 | 12.39 | |
| 10-10 | 0.74 | 0.44 | 0.58 | 0.59 | 0.12 | 20.89 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
| N | 10-6 | 6133.75 | 6226.00 | 6081.97 | 6147.21 | 59.57 | 0.97 |
| 10-7 | 617.10 | 621.28 | 598.72 | 612.37 | 9.80 | 1.60 | |
| 10-8 | 51.33 | 57.58 | 52.90 | 53.94 | 2.62 | 4.92 | |
| 10-9 | 5.33 | 5.69 | 4.40 | 5.14 | 0.54 | 10.57 | |
| 10-10 | 0.82 | 0.78 | 0.44 | 0.68 | 0.17 | 25.07 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
| E | 10-6 | 4350.43 | 4378.88 | 4334.00 | 4387.77 | 47.95 | 1.09 |
| 10-7 | 461.63 | 443.83 | 441.59 | 449.02 | 8.97 | 2.00 | |
| 10-8 | 45.01 | 50.60 | 43.13 | 46.25 | 3.17 | 6.86 | |
| 10-9 | 4.38 | 4.87 | 4.03 | 4.43 | 0.34 | 7.78 | |
| 10-10 | 1.32 | 1.69 | 1.32 | 1.44 | 0.17 | 12.08 | |
| 10-11 | 0 | 0.44 | 0 | 0.15 | 0.21 | 141.42 | |
| S | 10-6 | 5891.09 | 5839.83 | 5942.93 | 5891.28 | 42.09 | 0.71 |
| 10-7 | 591.21 | 558.87 | 593.71 | 581.26 | 15.87 | 2.73 | |
| 10-8 | 56.59 | 55.57 | 60.52 | 57.56 | 2.13 | 3.71 | |
| 10-9 | 5.06 | 5.87 | 5.92 | 5.62 | 0.39 | 7.02 | |
| 10-10 | 0.88 | 0.88 | 1.32 | 1.03 | 0.21 | 20.20 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |

图4 四重 ddPCR 灵敏度检测一维图 A:FAM 通道,ORF1ab 基因检测一维图;B:HEX 通道,N 基因检测一维图;C:ROX 通道,E 基因检测一维图;D:CY5 通道,S 基因检测一维图
Fig. 4 One-dimensional map of sensitivity detected by quadruple ddPCR A: FAM channel, one-dimensional map of ORF1ab gene detection; B: HEX channel, one-dimensional map of N-gene detection; C: ROX channel, one-dimensional map of E gene detection; D: CY5 channel, one-dimensional map of S gene detection
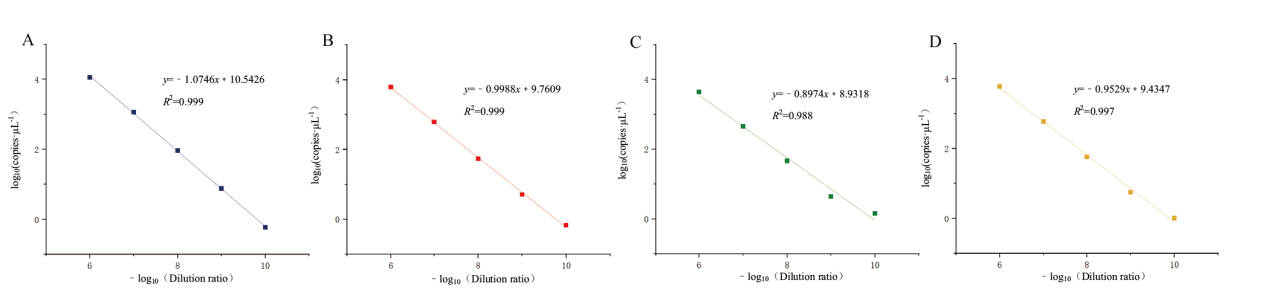
图5 四重 ddPCR 灵敏度检测线性关系拟合曲线图 A:ORF1ab 基因灵敏度检测线性关系拟合曲线图;B:N 基因灵敏度检测线性关系拟合曲线图;C:E 基因灵敏度检测线性关系拟合曲线图;D:S 基因灵敏度检测线性关系拟合曲线图
Fig. 5 Fitting curve of linear relationship for quadruple ddPCR sensitivity detection A: The fitting curve of the linear relationship of the sensitivity detection of ORF1ab gene. B: The fitting curve of the linear relationship of the sensitivity detection of N gene.C: The fitting curve of the linear relationship of the sensitivity detection of E gene. D: The fitting curve of the linear relationship of the sensitivity detection of S gene
| 样品名称No. of samples | 总微滴数Total number of droplets | ORF1ab/(Cp·μL-1) | N/(Cp·μL-1) | E/(Cp·μL-1) | S/(Cp·μL-1) |
|---|---|---|---|---|---|
| 样本1 | 20891 | 47.96 | 7480.95 | 335.57 | 99.29 |
| 样本2 | 21153 | 9.97 | 887.71 | 17.38 | 4.33 |
| 样本3 | 21372 | 4.70 | 219.61 | 9.71 | 6.45 |
| 样本4 | 20833 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本5 | 20536 | 28.03 | 6458.03 | 276.39 | 138.53 |
| 样本6 | 21393 | 14.59 | 912.05 | 21.85 | 11.59 |
| 样本7 | 21106 | 9.09 | 234.45 | 13.01 | 2.64 |
| 样本8 | 20712 | 248.53 | 7949.99 | 399.37 | 305.53 |
| 样本9 | 20793 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本10 | 21397 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本11 | 21368 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本12 | 21090 | 8.73 | 143.22 | 24.77 | 6.09 |
| 样本13 | 20675 | 5.57 | 112.42 | 19.40 | 2.20 |
| 样本14 | 20874 | 3.96 | 24.64 | 2.24 | 1.49 |
| 样本15 | 21704 | 93.39 | 6887.53 | 378.12 | 84.85 |
| 样本16 | 20380 | 38.92 | 780.27 | 357.50 | 21.19 |
| 样本17 | 21228 | 26.01 | 226.01 | 285.21 | 9.09 |
| 样本18 | 21104 | 29.11 | 269.19 | 14.39 | 24.35 |
| 样本19 | 20930 | 4.08 | 62.85 | 5.20 | 1.76 |
| 样本20 | 20678 | 118.65 | 10636.56 | 342.67 | 113.30 |
表3 20份样本多重微滴数字 PCR 拷贝数检测结果
Table 3 Copy number detection results for 20 samples by quadruple ddPCR
| 样品名称No. of samples | 总微滴数Total number of droplets | ORF1ab/(Cp·μL-1) | N/(Cp·μL-1) | E/(Cp·μL-1) | S/(Cp·μL-1) |
|---|---|---|---|---|---|
| 样本1 | 20891 | 47.96 | 7480.95 | 335.57 | 99.29 |
| 样本2 | 21153 | 9.97 | 887.71 | 17.38 | 4.33 |
| 样本3 | 21372 | 4.70 | 219.61 | 9.71 | 6.45 |
| 样本4 | 20833 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本5 | 20536 | 28.03 | 6458.03 | 276.39 | 138.53 |
| 样本6 | 21393 | 14.59 | 912.05 | 21.85 | 11.59 |
| 样本7 | 21106 | 9.09 | 234.45 | 13.01 | 2.64 |
| 样本8 | 20712 | 248.53 | 7949.99 | 399.37 | 305.53 |
| 样本9 | 20793 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本10 | 21397 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本11 | 21368 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本12 | 21090 | 8.73 | 143.22 | 24.77 | 6.09 |
| 样本13 | 20675 | 5.57 | 112.42 | 19.40 | 2.20 |
| 样本14 | 20874 | 3.96 | 24.64 | 2.24 | 1.49 |
| 样本15 | 21704 | 93.39 | 6887.53 | 378.12 | 84.85 |
| 样本16 | 20380 | 38.92 | 780.27 | 357.50 | 21.19 |
| 样本17 | 21228 | 26.01 | 226.01 | 285.21 | 9.09 |
| 样本18 | 21104 | 29.11 | 269.19 | 14.39 | 24.35 |
| 样本19 | 20930 | 4.08 | 62.85 | 5.20 | 1.76 |
| 样本20 | 20678 | 118.65 | 10636.56 | 342.67 | 113.30 |
| 样品名称 No. of samples | qPCR 检测结果(ct值)qPCR results | ||
|---|---|---|---|
| FAM(ORF1ab) | HEX(N) | ROX(E) | |
| 样本1 | 29.63 | 25.62 | 28.91 |
| 样本2 | 32.06 | 28.29 | 31.70 |
| 样本3 | 33.06 | 29.55 | 32.73 |
| 样本4 | NA | NA | NA |
| 样本5 | 30.63 | 26.95 | 29.49 |
| 样本6 | 31.61 | 28.17 | 30.55 |
| 样本7 | 32.75 | 29.41 | 31.78 |
| 样本8 | 27.36 | 25.29 | 25.46 |
| 样本9 | NA | NA | NA |
| 样本10 | NA | NA | NA |
| 样本11 | NA | NA | NA |
| 样本12 | 31.45 | 30.18 | 30.23 |
| 样本13 | 32.18 | 30.89 | 30.75 |
| 样本14 | 34.35 | 33.91 | 35.34 |
| 样本15 | 28.47 | 26.79 | 26.82 |
| 样本16 | 29.86 | 28.24 | 28.2 |
| 样本17 | 30.83 | 29.40 | 29.42 |
| 样本18 | 30.83 | 29.23 | 31.17 |
| 样本19 | 33.00 | 31.68 | 33.53 |
| 样本20 | 28.29 | 24.97 | 28.78 |
表4 20份样本荧光定量 PCR 检测结果
Table 4 Results of 20 samples via fluorescence quantitative PCR
| 样品名称 No. of samples | qPCR 检测结果(ct值)qPCR results | ||
|---|---|---|---|
| FAM(ORF1ab) | HEX(N) | ROX(E) | |
| 样本1 | 29.63 | 25.62 | 28.91 |
| 样本2 | 32.06 | 28.29 | 31.70 |
| 样本3 | 33.06 | 29.55 | 32.73 |
| 样本4 | NA | NA | NA |
| 样本5 | 30.63 | 26.95 | 29.49 |
| 样本6 | 31.61 | 28.17 | 30.55 |
| 样本7 | 32.75 | 29.41 | 31.78 |
| 样本8 | 27.36 | 25.29 | 25.46 |
| 样本9 | NA | NA | NA |
| 样本10 | NA | NA | NA |
| 样本11 | NA | NA | NA |
| 样本12 | 31.45 | 30.18 | 30.23 |
| 样本13 | 32.18 | 30.89 | 30.75 |
| 样本14 | 34.35 | 33.91 | 35.34 |
| 样本15 | 28.47 | 26.79 | 26.82 |
| 样本16 | 29.86 | 28.24 | 28.2 |
| 样本17 | 30.83 | 29.40 | 29.42 |
| 样本18 | 30.83 | 29.23 | 31.17 |
| 样本19 | 33.00 | 31.68 | 33.53 |
| 样本20 | 28.29 | 24.97 | 28.78 |
| [1] |
Shereen MA, Khan S, Kazmi A, et al. COVID-19 infection: emergence, transmission, and characteristics of human coronaviruses[J]. J Adv Res, 2020, 24: 91-98.
doi: 10.1016/j.jare.2020.03.005 URL |
| [2] |
李家俊, 郑潇, 盛杰, 等. 新型冠状病毒及其临床检测方法研究进展[J]. 生物技术通报, 2021, 37(4): 282-292.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0954 |
| Li JJ, Zheng X, Sheng J, et al. Novel coronavirus and research progress of related clinical detection methods[J]. Biotechnol Bull, 2021, 37(4): 282-292. | |
| [3] |
Andryukov BG, Besednova NN, Kuznetsova TA, et al. Laboratory-based resources for COVID-19 diagnostics: traditional tools and novel technologies. A perspective of personalized medicine[J]. J Pers Med, 2021, 11(1): 42.
doi: 10.3390/jpm11010042 URL |
| [4] |
Chan JFW, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan[J]. Emerg Microbes Infect, 2020, 9(1): 221-236.
doi: 10.1080/22221751.2020.1719902 URL |
| [5] |
Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin[J]. Nature, 2020, 579(7798): 270-273.
doi: 10.1038/s41586-020-2012-7 |
| [6] | Bal A, Destras G, Gaymard A, et al. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020[J]. Euro Surveill, 2021, 26(3): 2100008. |
| [7] |
Chen JH, Wang R, Wang ML, et al. Mutations strengthened SARS-CoV-2 infectivity[J]. J Mol Biol, 2020, 432(19): 5212-5226.
doi: S0022-2836(20)30456-3 pmid: 32710986 |
| [8] |
Dejnirattisai W, Shaw RH, Supasa P, et al. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum[J]. Lancet, 2022, 399(10321): 234-236.
doi: 10.1016/S0140-6736(21)02844-0 URL |
| [9] | Feder KA, Pearlowitz M, Goode A, et al. Linked clusters of SARS-CoV-2 variant B.1.351 - Maryland, january-february 2021[J]. Morb Mortal Wkly Rep, 2021, 70(17): 627-631. |
| [10] | Galloway SE, Paul P, MacCannell DR, et al. Emergence of SARS-CoV-2 B.1.1.7 lineage - United States, December 29, 2020-january 12, 2021[J]. Morb Mortal Wkly Rep, 2021, 70(3): 95-99. |
| [11] | 孙泽宇, 柴佳彤, 许建成. 新冠病毒变异株“奥密克戎”的研究进展[J]. 病毒学报, 2023, 39(2): 517-527. |
| Sun ZY, Chai JT, Xu JC. Research progress on the Omicron variant of SARS-CoV-2[J]. Chin J Virol, 2023, 39(2): 517-527. | |
| [12] |
Coutard B, Valle C, de Lamballerie X, et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade[J]. Antiviral Res, 2020, 176: 104742.
doi: 10.1016/j.antiviral.2020.104742 URL |
| [13] |
Benton DJ, Wrobel AG, Xu PQ, et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion[J]. Nature, 2020, 588(7837): 327-330.
doi: 10.1038/s41586-020-2772-0 |
| [14] |
Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR[J]. Nat Methods, 2013, 10(10): 1003-1005.
doi: 10.1038/nmeth.2633 pmid: 23995387 |
| [15] |
Li HY, Bai RL, Zhao ZY, et al. Application of droplet digital PCR to detect the pathogens of infectious diseases[J]. Biosci Rep, 2018, 38(6): BSR20181170.
doi: 10.1042/BSR20181170 URL |
| [16] | 王志彬, 罗庆, 李宝林, 等. 数字PCR技术检测感染者尿液中SARS-CoV-2的方法研究[J]. 西南医科大学学报, 2022, 45(4): 301-307. |
| Wang ZB, Luo Q, Li BL, et al. Study on the detection of SARS-CoV-2 in the urine of infected persons by droplet digital PCR[J]. J Southwest Med Univ, 2022, 45(4): 301-307. | |
| [17] | 杨朔鹏. 微滴式数字PCR用于生物与环境样本中SARS-CoV-2的高灵敏检测[D]. 石家庄: 河北医科大学, 2022. |
| Yang SP. Droplet digital PCR was used for the highly sensitive detection of SARS-CoV-2 in biological and environmental samples[D]. Shijiazhuang: Hebei Medical University, 2022. | |
| [18] |
Zhang Z, Wang N, Liu XF, et al. A novel, reverse transcription, droplet digital PCR assay for the combined, sensitive detection of severe acute respiratory syndrome coronavirus 2 with swine acute diarrhea syndrome coronavirus[J]. J AOAC Int, 2022, 105(5): 1437-1446.
doi: 10.1093/jaoacint/qsac039 pmid: 35377440 |
| [19] |
Collie S, Champion J, Moultrie H, et al. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa[J]. N Engl J Med, 2022, 386(5): 494-496.
doi: 10.1056/NEJMc2119270 URL |
| [20] |
Ren WL, Zhang Y, Rao JH, et al. Evolution of immune evasion and host range expansion by the SARS-CoV-2 B.1.1.529(Omicron)variant[J]. mBio, 2023, 14(2): e0041623.
doi: 10.1128/mbio.00416-23 URL |
| [21] |
Qu PK, Evans JP, Faraone JN, et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2[J]. Cell Host Microbe, 2023, 31(1): 9-17.e3.
doi: 10.1016/j.chom.2022.11.012 URL |
| [22] |
Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus(2019-nCoV)causing an outbreak of pneumonia[J]. Clin Chem, 2020, 66(4): 549-555.
doi: 10.1093/clinchem/hvaa029 URL |
| [23] | Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus(2019-nCoV)by real-time RT-PCR[J]. Euro Surveill, 2020, 25(3): 2000045. |
| [24] | 张稳健, 吕欣, 黄驰, 等. 胶体金免疫层析法检测新型冠状病毒IgM/IgG抗体的临床评价与应用[J]. 病毒学报, 2020, 36(3): 348-354. |
| Zhang WJ, Lyu X, Huang C, et al. Clinical evaluation and application of detection of IgM and IgG antibodies against SARS-CoV-2 using a colloidal gold immunochromatography assay[J]. Chin J Virol, 2020, 36(3): 348-354. | |
| [25] |
Cremonesi P, Cortimiglia C, Picozzi C, et al. Development of a droplet digital polymerase chain reaction for rapid and simultaneous identification of common foodborne pathogens in soft cheese[J]. Front Microbiol, 2016, 7: 1725.
pmid: 27840628 |
| [1] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [2] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [3] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [4] | 宋海娜, 吴心桐, 杨鲁豫, 耿喜宁, 张华敏, 宋小龙. 葱鳞葡萄孢菌诱导下韭菜RT-qPCR内参基因的筛选和验证[J]. 生物技术通报, 2023, 39(3): 101-115. |
| [5] | 穆德添, 万凌云, 章瑶, 韦树根, 陆英, 付金娥, 田艺, 潘丽梅, 唐其. 钩藤管家基因筛选及生物碱合成相关基因的表达分析[J]. 生物技术通报, 2023, 39(2): 126-138. |
| [6] | 李奕雅, 吴一凡, 丁能水, 范小萍, 陈凡. 荧光素酶辅助定量大肠杆菌破碎效果的方法[J]. 生物技术通报, 2023, 39(12): 90-98. |
| [7] | 李会杰, 董莲华, 陈桂芳, 刘思渊, 杨佳怡, 杨靖亚. 食品中椰毒假单胞菌微滴式数字PCR定量检测方法的建立[J]. 生物技术通报, 2023, 39(1): 127-136. |
| [8] | 王祥锟, 宋学宏, 刘金龙, 郭培红, 庄晓峰, 韦良孟, 周凡, 张树宇, 高攀攀, 魏凯. 新型冠状病毒亚单位疫苗研制及其高效免疫增强剂的筛选[J]. 生物技术通报, 2023, 39(1): 305-314. |
| [9] | 曹英芳, 赵新, 刘双, 李瑞环, 刘娜, 徐石勇, 高芳瑞, 马卉, 兰青阔, 檀建新, 王永. 抗除草剂大豆GE-J12实时荧光定量PCR检测方法的建立[J]. 生物技术通报, 2022, 38(7): 146-152. |
| [10] | 刘晓玫, 王东鑫, 张春, 魏双施. AAV介导的RNAi对SARS-CoV-2 S基因表达的抑制作用[J]. 生物技术通报, 2022, 38(3): 188-193. |
| [11] | 粟元, 朱龙佼, 曹继娟, 刘建龙, 许文涛. 基于大肠埃希菌 O157∶H7的荧光定量冻干检测试剂盒的研制[J]. 生物技术通报, 2022, 38(3): 264-275. |
| [12] | 兰欣悦, 刘宁宁, 朱龙佼, 陈旭, 褚华硕, 李相阳, 段诺, 许文涛. 四环素双价适配体非酶免标记传感器[J]. 生物技术通报, 2022, 38(3): 276-284. |
| [13] | 苗玉娇, 朱龙佼, 许文涛. 质谱成像新基质及其在分析生物样本方面的研究进展[J]. 生物技术通报, 2022, 38(12): 156-167. |
| [14] | 蒋旭东, 刘宇, 邬建飞, 胡双阁, 卢建远, 字向东. 牦牛FGG组织表达与雌性生殖器官中定位分析[J]. 生物技术通报, 2022, 38(11): 286-294. |
| [15] | 徐圆圆, 赵国春, 郝颖颖, 翁学煌, 陈仲, 贾黎明. 无患子RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2022, 38(10): 80-89. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||