








生物技术通报 ›› 2024, Vol. 40 ›› Issue (6): 319-329.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1228
王梦帆1( ), 赵子玉1, 王春光1, 刘廷玉2, 陈曦1, 张铁1(
), 赵子玉1, 王春光1, 刘廷玉2, 陈曦1, 张铁1( )
)
收稿日期:2024-01-02
出版日期:2024-06-26
发布日期:2024-06-24
通讯作者:
张铁,男,硕士,教授,研究方向:中兽药防治畜禽疾病;E-mail: zhangtie1998@163.com作者简介:王梦帆,女,硕士研究生,研究方向:细菌耐药性;E-mail: mengfan202312@163.com赵子玉为本文共同第一作者
基金资助:
WANG Meng-fan1( ), ZHAO Zi-yu1, WANG Chun-guang1, LIU Ting-yu2, CHEN Xi1, ZHANG Tie1(
), ZHAO Zi-yu1, WANG Chun-guang1, LIU Ting-yu2, CHEN Xi1, ZHANG Tie1( )
)
Received:2024-01-02
Published:2024-06-26
Online:2024-06-24
摘要:
【目的】 开发针对头孢噻肟-水解酶-14(CTX-M-14)型超广谱β-内酰胺酶(extended-spectrum β-lactamase,ESBLs)的抑制剂,用以缓解细菌耐药带来的严重危害。【方法】 使用DiscoveryStudio Visualizer(DS Visualizer)构建基于受体结构的药效团模型(structure-based pharmacophore, SBP)和基于配体共同特征的定性药效团模型(common feature pharmacophore generation, HIPHOP),并将验证后的模型作为查询条件对ZINC数据库进行以CTX-M-14蛋白为靶标的虚拟筛选,得到拟合分数良好的中药单体成分甘草酸(glycyrrhizic acid,GL),对其进行相关作用力分析、分子动力学模拟和结合自由能计算,分析甘草酸与CTX-M-14蛋白的结合模式、结合能力及稳定性;最后通过联合抑菌试验及酶动力学试验考察甘草酸的抗菌增敏活性、抑酶作用及抑酶方式。【结果】 中药单体成分甘草酸主要与CTX-M-14蛋白活性中心多个氨基酸残基形成氢键和范德华作用力,两者的对接分数与结合自由能分别为-10 kcal/mol及-22.06 kcal/mol;甘草酸与头孢噻肟钠联用呈协同作用(FICI≤0.5);甘草酸可竞争性抑制β-内酰胺酶对底物抗生素的水解作用,对头孢噻肟钠的抑酶保护率可达58.53%,与克拉维酸接近(60.98%)。【结论】 中药单体甘草酸可与CTX-M-14蛋白稳定结合,通过竞争性抑制β-内酰胺酶对底物抗生素的水解作用,提高多重耐药大肠杆菌E320和重组蛋白阳性菌BL-21对头孢噻肟钠的敏感性,实现抗生素的减量增效。
王梦帆, 赵子玉, 王春光, 刘廷玉, 陈曦, 张铁. 基于药效团模型筛选CTX-M-14型超广谱β-内酰胺酶抑制剂[J]. 生物技术通报, 2024, 40(6): 319-329.
WANG Meng-fan, ZHAO Zi-yu, WANG Chun-guang, LIU Ting-yu, CHEN Xi, ZHANG Tie. Screening of CTX-M-14-type Ultra-broad-spectrum β-lactamase Inhibitors Based on Pharmacophore Modelling[J]. Biotechnology Bulletin, 2024, 40(6): 319-329.
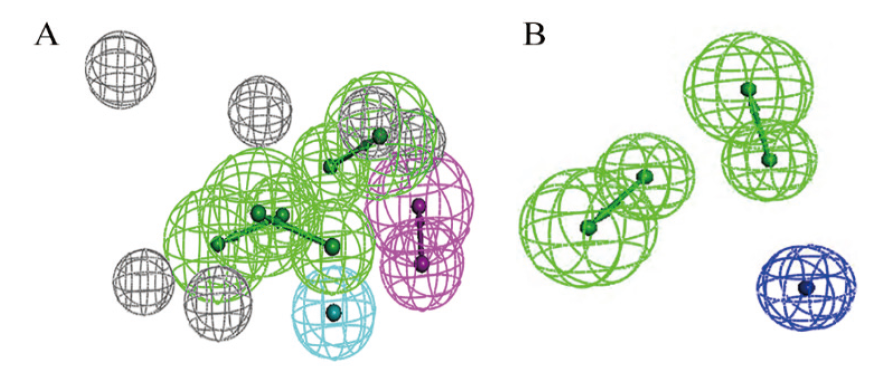
图2 基于CTX-M-14蛋白受体(A)及配体共同特征(B)药效团模型 绿色箭头和绿色网状球形,氢键受体;紫色箭头和紫色网状球形,氢键供体;浅蓝色圆球和蓝色网状球形,疏水作用力;深蓝色圆球和深蓝色网状球形,负电荷中心;灰色网状球形,排除体积
Fig. 2 Pharmacophore modelling based on CTX-M-14 protein receptor(A)and ligand co-features(B) Green arrow and green reticulated sphere, hydrogen bond acceptor; purple arrow and purple reticulated sphere, hydrogen bond donor; light blue orb and blue reticulated sphere, hydrophobic forces; dark blue orb and dark blue reticulated sphere, negative charge centre; grey reticulated sphere, excluded volume

图4 SBP模型(A)和HIPHOP模型(B)与测试集化合物拟合示意图,甘草酸与SBP模型(C)和HIPHOP模型(D)拟合示意图
Fig. 4 Schematic of SBP model (A) and HIPHOP model (B) fitted to the test set of compounds and glycyrrhizic acid fitted to the SBP model (C) and HIPHOP model (D)
| ZINC ID | 名称Name | 拟合值FitValue | |
|---|---|---|---|
| SBP | HIPHOP | ||
| ZINC901518 | 茶渍酸Lecanoric acid | 2.6156 | 2.32539 |
| ZINC96015174 | 甘草酸Glycyrrhizin | 2.03995 | 2.44671 |
| ZINC4164596 | 富马前冰岛酸Fumarprotocetraric acid | 1.25871 | 2.61666 |
表1 SBP和HIPHOP模型同时命中的小分子化合物及其FitValue
Table 1 Small molecule compounds hit simultaneously by SBP and HIPHOP models and their FitValues
| ZINC ID | 名称Name | 拟合值FitValue | |
|---|---|---|---|
| SBP | HIPHOP | ||
| ZINC901518 | 茶渍酸Lecanoric acid | 2.6156 | 2.32539 |
| ZINC96015174 | 甘草酸Glycyrrhizin | 2.03995 | 2.44671 |
| ZINC4164596 | 富马前冰岛酸Fumarprotocetraric acid | 1.25871 | 2.61666 |
| ZINC ID | 名称 Name | 结合自由能Affinity*/ (kcal·mol-1) | 均方根偏差/RMSD | |
|---|---|---|---|---|
| 最小值l.b. | 最大值u.b. | |||
| ZINC901518 | 茶渍酸Lecanoric acid | -8.3 | 0 | 0 |
| ZINC96015174 | 甘草酸Glycyrrhizin | -10.0 | 0 | 0 |
| ZINC4164596 | 富马前冰岛酸Fumarprotocetraric acid | -9.4 | 0 | 0 |
表2 SBP模型和HIPHOP模型同时命中的候选化合物与CTX-M-14蛋白对接分数
Table 2 Docking scores of candidate compounds and CTX-M-14 proteins hit simultaneously by the SBP model and HIPHOP model
| ZINC ID | 名称 Name | 结合自由能Affinity*/ (kcal·mol-1) | 均方根偏差/RMSD | |
|---|---|---|---|---|
| 最小值l.b. | 最大值u.b. | |||
| ZINC901518 | 茶渍酸Lecanoric acid | -8.3 | 0 | 0 |
| ZINC96015174 | 甘草酸Glycyrrhizin | -10.0 | 0 | 0 |
| ZINC4164596 | 富马前冰岛酸Fumarprotocetraric acid | -9.4 | 0 | 0 |

图5 甘草酸与CTX-M-14蛋白分子对接示意图 绿色、粉色和黄色圆形,氨基酸残基;绿色、粉色和黄色虚线,相互作用力;灰色线形,甘草酸配体;灰色粗棒,甘草酸配体;灰色细棒,氨基酸残基;黑色字体,氨基酸残基名称
Fig. 5 Schematic diagram of molecular docking of glycyrrhizic acid with CTX-M-14 protein+ Green, pink and yellow circles, amino acid residues; green, pink and yellow dashed lines, interacting forces; grey lines, glycyrrhizic acid ligands; grey thick rods, glycyrrhizic acid ligands; grey thin rods, amino acid residues; black lettering, amino acid residue names

图6 甘草酸与CTX-M-14蛋白复合物分子动力学模拟的RMSD(A)和RMSF(B)
Fig. 6 RMSD(A)and RMSF(B)of molecular dynamics simulations of glycyrrhizic acid complexed with CTX-M-14 protein
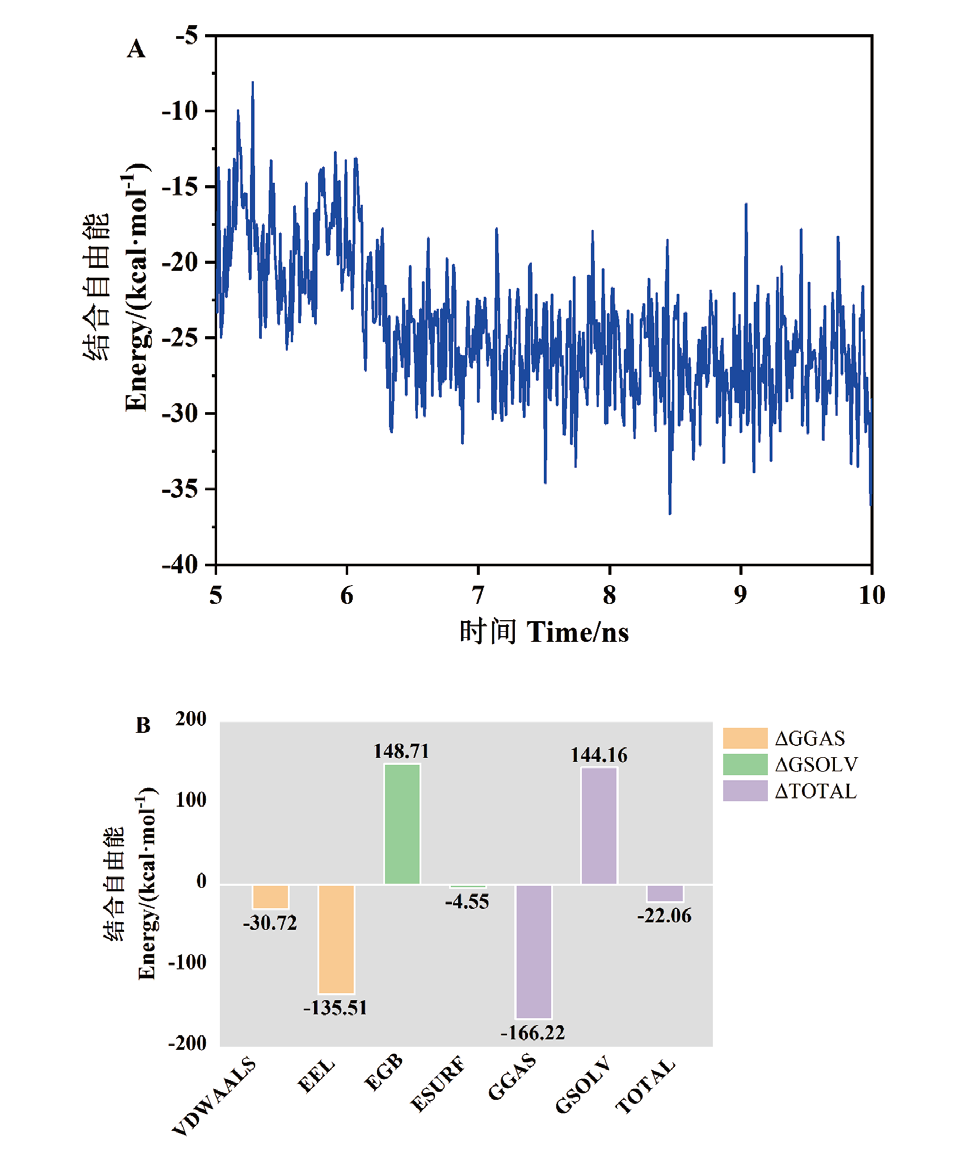
图7 甘草酸与CTX-M-14蛋白复合物动力学模拟的平衡轨迹采样(A)和结合自由能分解(B)
Fig. 7 Equilibrium trajectory sampling(A)and binding free energy decomposition(B)for kinetic simulations of glycyrrhetinic acid complexes with CTX-M-14 protein
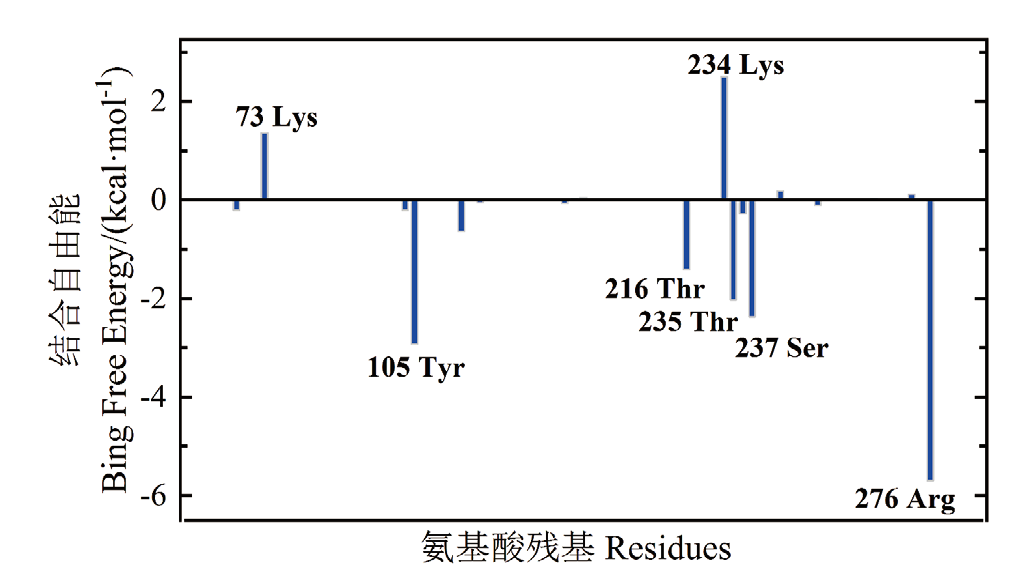
图8 甘草酸与CTX-M-14蛋白相互作用中关键氨基酸残基及其结合自由能分解
Fig. 8 Key amino acid residues and their binding free energy decomposition in the interaction between glycyrrhetinic acid and CTX-M-14 protein
| 药物Drug/(mg·mL-1) | E320 | 蛋白重组阳性菌BL-21 Protein recombinant positive bacterial strain BL-21 |
|---|---|---|
| 甘草酸Glycyrrhizic acid | >25 | >25 |
| 苯唑西林钠Benzoxacillin sodium | >256(R) | 64(R) |
| 氨苄西林钠Ampicillin sodium | >256(R) | 64(R) |
| 头孢唑林钠Cefazolin sodium | >256(R) | 16(R) |
| 头孢他啶Ceftazidime | >256(R) | 8(I) |
| 头孢呋辛Cefuroxime | 4(S) | 16(I) |
| 头孢曲松钠Ceftriaxone sodium | >256(R) | 32(R) |
| 头孢噻肟钠Cefotaxime sodium | 1024(R) | >1024(R) |
表3 甘草酸和抗菌药物对大肠杆菌的 MIC
Table 3 MIC of glycyrrhetinic acid and antibiotics against Escherichia coli
| 药物Drug/(mg·mL-1) | E320 | 蛋白重组阳性菌BL-21 Protein recombinant positive bacterial strain BL-21 |
|---|---|---|
| 甘草酸Glycyrrhizic acid | >25 | >25 |
| 苯唑西林钠Benzoxacillin sodium | >256(R) | 64(R) |
| 氨苄西林钠Ampicillin sodium | >256(R) | 64(R) |
| 头孢唑林钠Cefazolin sodium | >256(R) | 16(R) |
| 头孢他啶Ceftazidime | >256(R) | 8(I) |
| 头孢呋辛Cefuroxime | 4(S) | 16(I) |
| 头孢曲松钠Ceftriaxone sodium | >256(R) | 32(R) |
| 头孢噻肟钠Cefotaxime sodium | 1024(R) | >1024(R) |
| 菌株Strain | 药物Drug | 单药MIC MIC of single drug | 联合用药MIC MIC of combined drugs | FICI | 结果判定Result determination |
|---|---|---|---|---|---|
| E320 | 甘草酸Glycyrrhizic acid(mg·mL-1) | >25 | 3.125 | <0.25 | 协同作用 Synergy |
| 头孢噻肟钠Cefotaxime(μg·mL-1) | 1 024 | 128 | |||
| CTX-M-14蛋白重组阳性菌CTX-M-14 Protein recombinant positive bacteria | 甘草酸Glycyrrhizic acid(mg·mL-1) | >25 | 3.125 | <0.375 | 协同作用 Synergy |
| 头孢噻肟钠Cefotaxime(μg·mL-1) | 1 024 | 256 | |||
| 蛋白重组阴性对照菌Protein recombinant negative control bacteria | 甘草酸Glycyrrhizic acid(mg·mL-1) | >25 | 1.5625 | <1.0625 | 无关作用 Unrelated effects |
| 头孢噻肟钠Cefotaxime(μg·mL-1) | 8 | 8 |
表4 甘草酸与头孢噻肟钠联合抑菌试验
Table 4 Combination antibacterial test of glycyrrhetinic acid and cefotaxime sodium
| 菌株Strain | 药物Drug | 单药MIC MIC of single drug | 联合用药MIC MIC of combined drugs | FICI | 结果判定Result determination |
|---|---|---|---|---|---|
| E320 | 甘草酸Glycyrrhizic acid(mg·mL-1) | >25 | 3.125 | <0.25 | 协同作用 Synergy |
| 头孢噻肟钠Cefotaxime(μg·mL-1) | 1 024 | 128 | |||
| CTX-M-14蛋白重组阳性菌CTX-M-14 Protein recombinant positive bacteria | 甘草酸Glycyrrhizic acid(mg·mL-1) | >25 | 3.125 | <0.375 | 协同作用 Synergy |
| 头孢噻肟钠Cefotaxime(μg·mL-1) | 1 024 | 256 | |||
| 蛋白重组阴性对照菌Protein recombinant negative control bacteria | 甘草酸Glycyrrhizic acid(mg·mL-1) | >25 | 1.5625 | <1.0625 | 无关作用 Unrelated effects |
| 头孢噻肟钠Cefotaxime(μg·mL-1) | 8 | 8 |
| 组别Group | Km | Vmax | Ki | Km/Vmax |
|---|---|---|---|---|
| 空白对照组Blank control group | 7.13 | 21.33 | - | 0.33 |
| 溶液对照组Solution control group | 7.11 | 20.97 | - | 0.34 |
| 克拉维酸组Clavulanic acid group | 8.76 | 17.79 | 3.78 | 0.50 |
| 甘草酸Glycyrrhetinic acid group | 8.39 | 18.03 | 9.69 | 0.47 |
表5 CTX-M-14蛋白酶动力学参数
Table 5 CTX-M-14 protease kinetic parameters
| 组别Group | Km | Vmax | Ki | Km/Vmax |
|---|---|---|---|---|
| 空白对照组Blank control group | 7.13 | 21.33 | - | 0.33 |
| 溶液对照组Solution control group | 7.11 | 20.97 | - | 0.34 |
| 克拉维酸组Clavulanic acid group | 8.76 | 17.79 | 3.78 | 0.50 |
| 甘草酸Glycyrrhetinic acid group | 8.39 | 18.03 | 9.69 | 0.47 |
| 组别Group | 初始OD值 Initial OD value | OD值 OD value | OD值变化量 OD value variation | 抑酶保护率 Enzyme-inhibition protection rate/% |
|---|---|---|---|---|
| 空白对照组Blank control group | 1.15±0.035 | 0.33±0.015 | 0.82±0.026 | - |
| 溶液对照组Solution control group | 1.14±0.020 | 0.32±0.025 | 0.81±0.015 | - |
| 克拉维酸组Clavulanic acid group | 1.19±0.025 | 0.87±0.020 | 0.32±0.020* | 60.98 |
| 甘草酸组Glycyrrhetinic acid group | 1.31±0.017 | 0.98±0.010 | 0.34±0.022* | 58.53 |
表6 甘草酸对头孢噻肟钠的抑酶保护率
Table 6 Enzyme-inhibition protection rate of glycyrrhetinic acid against cefotaxime sodium
| 组别Group | 初始OD值 Initial OD value | OD值 OD value | OD值变化量 OD value variation | 抑酶保护率 Enzyme-inhibition protection rate/% |
|---|---|---|---|---|
| 空白对照组Blank control group | 1.15±0.035 | 0.33±0.015 | 0.82±0.026 | - |
| 溶液对照组Solution control group | 1.14±0.020 | 0.32±0.025 | 0.81±0.015 | - |
| 克拉维酸组Clavulanic acid group | 1.19±0.025 | 0.87±0.020 | 0.32±0.020* | 60.98 |
| 甘草酸组Glycyrrhetinic acid group | 1.31±0.017 | 0.98±0.010 | 0.34±0.022* | 58.53 |
| [1] | Husna A, Rahman MM, Badruzzaman ATM, et al. Extended-spectrum β-lactamases(ESBL): challenges and opportunities[J]. Biomedicines, 2023, 11(11): 2937. |
| [2] |
Collaborators AR. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis[J]. Lancet, 2022, 399(10325): 629-655.
doi: 10.1016/S0140-6736(21)02724-0 pmid: 35065702 |
| [3] | Ramatla T, Mafokwane T, Lekota K, et al. “One Health” perspective on prevalence of co-existing extended-spectrum β-lactamase(ESBL)producing Escherichia coli and Klebsiella pneumoniae: a comprehensive systematic review and meta-analysis[J]. Ann Clin Microbiol Antimicrob, 2023, 22(1): 88. |
| [4] |
Peirano G, Pitout JDD. Extended-spectrum β-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options[J]. Drugs, 2019, 79(14): 1529-1541.
doi: 10.1007/s40265-019-01180-3 pmid: 31407238 |
| [5] | Castanheira M, Simner PJ, Bradford PA. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection[J]. JAC Antimicrob Resist, 2021, 3(3): dlab092. |
| [6] |
Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors[J]. Clin Microbiol Rev, 2010, 23(1): 160-201.
doi: 10.1128/CMR.00037-09 pmid: 20065329 |
| [7] |
Bush K, Bradford PA. Interplay between β-lactamases and new β-lactamase inhibitors[J]. Nat Rev Microbiol, 2019, 17(5): 295-306.
doi: 10.1038/s41579-019-0159-8 pmid: 30837684 |
| [8] |
Wang YH, Wang J, Wang R, et al. Resistance to ceftazidime-avibactam and underlying mechanisms[J]. J Glob Antimicrob Resist, 2020, 22: 18-27.
doi: S2213-7165(19)30323-6 pmid: 31863899 |
| [9] | 曹敏. 天然β-内酰胺酶抑制剂的筛选研究[D]. 贵阳: 贵州大学, 2016. |
| Cao M. The research on inhibitors screened from natural medicine monomers[D]. Guiyang: Guizhou University, 2016. | |
| [10] | 刘立新, 高月林, 王朝兴, 等. 黄酮化合物对金黄葡萄球菌β-内酰胺酶活性影响[J]. 东北农业大学学报, 2013, 44(3): 119-122. |
| Liu LX, Gao YL, Wang CX, et al. Effect of the flavonoids on the activity of β-lactamase from Staphylococcus aureus[J]. J Northeast Agric Univ, 2013, 44(3): 119-122. | |
| [11] | 李敏敏. 黄连与环丙沙星联用对鸡源性沙门菌生物被膜的影响[D]. 哈尔滨: 东北农业大学, 2016. |
| Li MM. Effects of rhizoma coptidis in combination with ciprofloxacin on Salmonella biofilm that isolated from chickens[D]. Harbin: Northeast Agricultural University, 2016. | |
| [12] | Mgbeahuruike EE, Stålnacke M, Vuorela H, et al. Antimicrobial and synergistic effects of commercial piperine and piperlongumine in combination with conventional antimicrobials[J]. Antibiotics, 2019, 8(2): 55. |
| [13] |
朱浩, 张严伟, 刘润, 等. 抗生素佐剂与抗生素联用的抑菌作用研究进展[J]. 生物技术通报, 2022, 38(6): 66-73.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0027 |
|
Zhu H, Zhang YW, Liu R, et al. Research progress in antibiotic adjuvant and antibiotics in antibacterial aspects[J]. Biotechnol Bull, 2022, 38(6): 66-73.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0027 |
|
| [14] |
Mysinger MM, Carchia M, Irwin JJ, et al. Directory of useful decoys, enhanced(DUD-E): better ligands and decoys for better benchmarking[J]. J Med Chem, 2012, 55(14): 6582-6594.
doi: 10.1021/jm300687e pmid: 22716043 |
| [15] | 王亚铃, 姚明丽, 钱晨亮, 等. 基于药效团和分子对接筛选D-丙氨酰-D-丙氨酸连接酶抑制剂[J]. 江苏海洋大学学报: 自然科学版, 2023, 32(2): 53-59. |
| Wang YL, Yao ML, Qian CL, et al. High-throughput virtual screening of D-alanyl-D-alanine ligase inhibitors based on pharmacophore and molecular docking[J]. J Jiangsu Ocean Univ Nat Sci Ed, 2023, 32(2): 53-59. | |
| [16] | 姜星, 靳文珂, 李自祥, 等. CPS1小分子抑制剂的筛选及其抗结直肠癌的机制研究[J]. 药学学报, 2022, 57(9): 2671-2681. |
| Jiang X, Jin WK, Li ZX, et al. Discovery of a small-molecule inhibitor of carbamoyl phosphate synthase 1 and its anti-colorectal cancer mechanism[J]. Acta Pharm Sin, 2022, 57(9): 2671-2681. | |
| [17] |
赵子玉, 王春光, 吕建存, 等. 超广谱β-内酰胺酶CTX-M-14中药抑制剂的筛选及芸香苷抑酶作用研究[J]. 生物技术通报, 2022, 38(6): 235-244.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1105 |
| Zhao ZY, Wang CG, Lv JC, et al. Screening of β-lactamase CTX-M-14 Chinese medicine inhibitor and enzyme inhibition of rutin[J]. Biotechnol Bull, 2022, 38(6): 235-244. | |
| [18] | 张莹莹. 基于结构的NSD2小分子抑制剂筛选及初步活性验证[D]. 郑州: 河南工业大学, 2023. |
| Zhang YY. Structure-based drug design and biological activity evaluation of methyltransferase NSD2[D]. Zhengzhou: Henan University of Technology, 2023. | |
| [19] | Giordano D, Biancaniello C, Argenio MA, et al. Drug design by pharmacophore and virtual screening approach[J]. Pharmaceuticals, 2022, 15(5): 646. |
| [20] | 郑晓杰. Src和Abl单双靶点抑制剂的虚拟筛选及分子动力学研究[D]. 广州: 广东药科大学, 2019. |
| Zheng XJ. Virtual screening and molecular dynamics study of src and abl single and double target inhibitors[D]. Guangzhou: Guangdong Pharmaceutical University, 2019. | |
| [21] | 刘璐. 甘草酸作为活性“药用辅料” 可行性实验研究[D]. 青岛: 青岛科技大学, 2023. |
| Liu L. Feasibility study of glycyrrhizin acid as an active‘Pharmaceutical excipient’[D]. Qingdao: Qingdao University of Science & Technology, 2023. | |
| [22] |
Paudel YN, Angelopoulou E, Semple B, et al. Potential neuroprotective effect of the HMGB1 inhibitor glycyrrhizin in neurological disorders[J]. ACS Chem Neurosci, 2020, 11(4): 485-500.
doi: 10.1021/acschemneuro.9b00640 pmid: 31972087 |
| [23] | Huan CC, Xu Y, Zhang W, et al. Research progress on the antiviral activity of glycyrrhizin and its derivatives in liquorice[J]. Front Pharmacol, 2021, 12: 680674. |
| [24] | Alagawany M, Elnesr SS, Farag MR, et al. Use of licorice(Glycyr-rhiza glabra)herb as a feed additive in poultry: current knowledge and prospects[J]. Animals, 2019, 9(8): 536. |
| [25] | Perbandt M, Werner N, Prester A, et al. Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as β-lactamase inhibitors[J]. Sci Rep, 2022, 12(1): 5510. |
| [26] | Pemberton OA, Tsivkovski R, Totrov M, et al. Structural basis and binding kinetics of vaborbactam in class A β-lactamase inhibition[J]. Antimicrob Agents Chemother, 2020, 64(10): e00398-20. |
| [27] | Ahmadvand P, Avillan JJ, Lewis JA, et al. Characterization of interactions between CTX-M-15 and clavulanic acid, desfuroylceftiofur, ceftiofur, ampicillin, and nitrocefin[J]. Int J Mol Sci, 2022, 23(9): 5229. |
| [28] | 梁蕾蕾. 细胞色素P450 4F2与α-生育酚结合机制的分子动力学模拟研究[D]. 长春: 吉林大学, 2023. |
| Liang LL. Investigation of the binding mechanism between α-TOH and CYP4F2 using molecular dynamics simulations[D]. Changchun: Jilin University, 2023. | |
| [29] | 王瑞歌. HIV-1蛋白酶与抑制剂相互作用的分子动力学模拟研究[D]. 长春: 吉林大学, 2021. |
| Wang RG. Study on the interaction between HIV-1 proetase and inhibitors using molecular dynamics simulations[D]. Changchun: Jilin University, 2021. | |
| [30] |
Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes[J]. Antimicrob Agents Chemother, 2004, 48(1): 1-14.
doi: 10.1128/AAC.48.1.1-14.2004 pmid: 14693512 |
| [31] | Readman JB, Acman M, Hamawandi A, et al. Cefotaxime/sulbactam plus gentamicin as a potential carbapenem- and amikacin-sparing first-line combination for neonatal sepsis in high ESBL prevalence settings[J]. J Antimicrob Chemother, 2023, 78(8): 1882-1890. |
| [1] | 史京辉, 陈文慧, 陆坤, 郑婷婷, 任志远, 鲍国庆, 王敏, 骆健美. 定点饱和突变提高赭曲霉11α羟化酶的催化性能[J]. 生物技术通报, 2024, 40(1): 322-331. |
| [2] | 周定定, 李辉虎, 汤兴涌, 余发新, 孔丹宇, 刘毅. 甘草酸和甘草苷生物合成与调控的研究进展[J]. 生物技术通报, 2023, 39(5): 44-53. |
| [3] | 赵子玉, 王春光, 吕建存, 李继开, 张铁. 超广谱β-内酰胺酶CTX-M-14中药抑制剂的筛选及芸香苷抑酶作用研究[J]. 生物技术通报, 2022, 38(6): 235-244. |
| [4] | 孙熙麟, 蒋振彦, 刘志屹, 戴璐, 孙非, 黄伟. 氨基酸定点突变提高灵芝蛋白LZ-8热稳定性的研究[J]. 生物技术通报, 2020, 36(1): 23-28. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||