








生物技术通报 ›› 2024, Vol. 40 ›› Issue (9): 11-19.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0520
• 薯类作物生物技术专题(专题主编:徐建飞,尚轶) • 上一篇 下一篇
童玮婧1,2( ), 罗数1, 陆新露1, 沈建福3, 陆柏益3, 李开绵4, 马秋香1(
), 罗数1, 陆新露1, 沈建福3, 陆柏益3, 李开绵4, 马秋香1( ), 张鹏1,2(
), 张鹏1,2( )
)
收稿日期:2024-05-30
出版日期:2024-09-26
发布日期:2024-10-12
通讯作者:
马秋香,女,副研究员,研究方向:薯类分子育种;E-mail: qxma@cemps.ac.cn;作者简介:童玮婧,女,博士研究生,研究方向:薯类分子育种;E-mail: tongweijing@cemps.ac.cn
基金资助:
TONG Wei-jing1,2( ), LUO Shu1, LU Xin-lu1, SHEN Jian-fu3, LU Bai-yi3, LI Kai-mian4, MA Qiu-xiang1(
), LUO Shu1, LU Xin-lu1, SHEN Jian-fu3, LU Bai-yi3, LI Kai-mian4, MA Qiu-xiang1( ), ZHANG Peng1,2(
), ZHANG Peng1,2( )
)
Received:2024-05-30
Published:2024-09-26
Online:2024-10-12
摘要:
【目的】木薯(Manihot esculenta Crantz)中含有潜在毒性的生氰糖苷,其食用安全性受到影响且导致加工成本增加。因此,利用生物技术开展低生氰糖苷木薯的培育具有重要意义。【方法】利用CRISPR/Cas9技术对木薯醇氰酶基因MeHNL进行了编辑。该基因编码催化生氰糖苷分解的α-羟基腈裂解酶,编辑靶点位于第1个外显子上,通过农杆菌介导的稳定转化获得了27株阳性植株。【结果】测序分析显示,其中26个株系被编辑,编辑效率高达96.3%。编辑类型主要包括碱基的插入和缺失,少数为碱基替换和大片段缺失。氰化物检测试剂盒染色和HPLC测定分析表明,编辑株系中的氢氰酸和生氰糖苷含量均显著降低。与非编辑植株相比,编辑植株的叶片细长,暗示了MeHNL可能对木薯的生长发育产生影响。【结论】利用CRISPR/Cas9技术获得了低氰化物的木薯种质,为开展生氰糖苷代谢影响木薯生长发育的研究提供了材料。
童玮婧, 罗数, 陆新露, 沈建福, 陆柏益, 李开绵, 马秋香, 张鹏. CRISPR/Cas9编辑MeHNL基因创制低生氰糖苷木薯[J]. 生物技术通报, 2024, 40(9): 11-19.
TONG Wei-jing, LUO Shu, LU Xin-lu, SHEN Jian-fu, LU Bai-yi, LI Kai-mian, MA Qiu-xiang, ZHANG Peng. CRISPR/Cas9 Editing MeHNL Gene to Generate Cassava Plants with Low Cyanogenic Glycoside[J]. Biotechnology Bulletin, 2024, 40(9): 11-19.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Use |
|---|---|---|
| target-Fp | GATTGCTTGGAGAAACTCCCTCAAG | Target gene |
| target-Rp | AAACCTTGAGGGAGTTTCTCCAAGC | |
| M13-Fp | TGTAAAACGACGGCCAGT | Identification of positive clones |
| Hpt II-Fp | TTCTACACAGCCATCGGTCC | Identification of positive transgenic plants |
| Hpt II-Rp | CCCATGTGTATCACTGGCAA | |
| HNLgRNA-Fp | GCACCACTCACAGAAAAATCCAAAG | PCR amplification of target region |
| HNLgRNA-Rp | ATGTGGCTTAATTAGAATACCGTCT |
表1 实验中所用引物
Table 1 Primers used in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Use |
|---|---|---|
| target-Fp | GATTGCTTGGAGAAACTCCCTCAAG | Target gene |
| target-Rp | AAACCTTGAGGGAGTTTCTCCAAGC | |
| M13-Fp | TGTAAAACGACGGCCAGT | Identification of positive clones |
| Hpt II-Fp | TTCTACACAGCCATCGGTCC | Identification of positive transgenic plants |
| Hpt II-Rp | CCCATGTGTATCACTGGCAA | |
| HNLgRNA-Fp | GCACCACTCACAGAAAAATCCAAAG | PCR amplification of target region |
| HNLgRNA-Rp | ATGTGGCTTAATTAGAATACCGTCT |
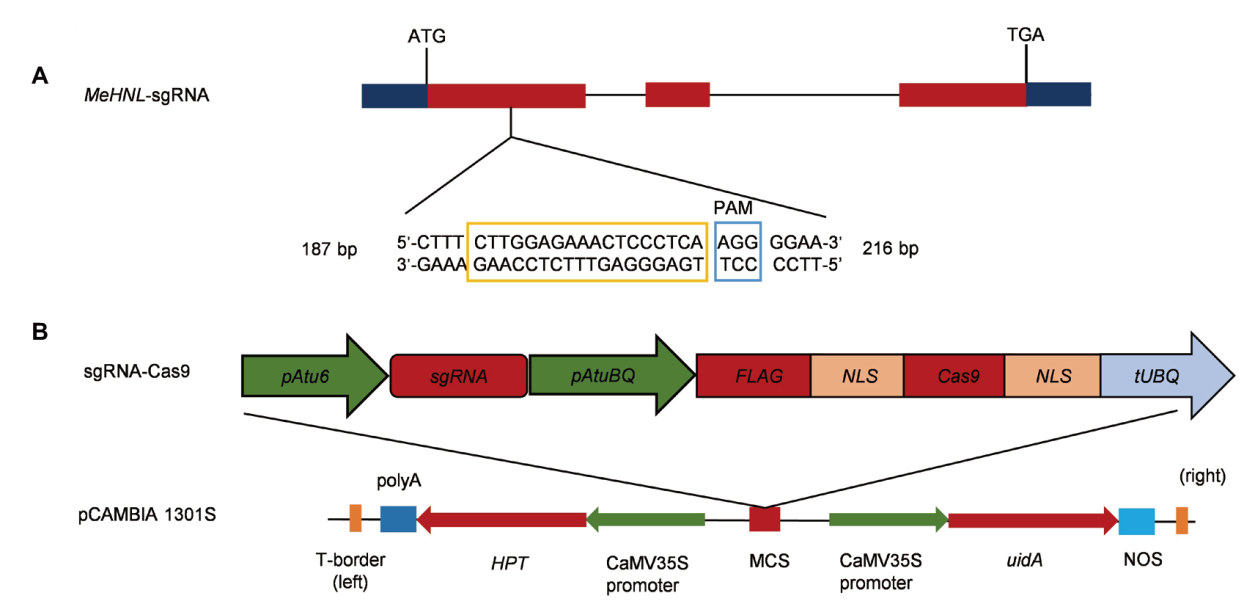
图1 CRISPR/Cas9编辑载体示意图 A:橘色方框内的核苷酸序列表示MeHNL基因编辑的靶标位点,蓝色方框内的核苷酸序列为PAM(Protospacer adjacent motif)位点;B:CRISPR/Cas9载体骨架
Fig. 1 Schematic diagram of CRISPR/Cas9 editing vector A: The nucleotide sequences in orange box indicates the target site of MeHNL gene, the nucleotide sequences in blue box indicates the PAM site. B: The skeleton of CRISPR/Cas9 expression vector
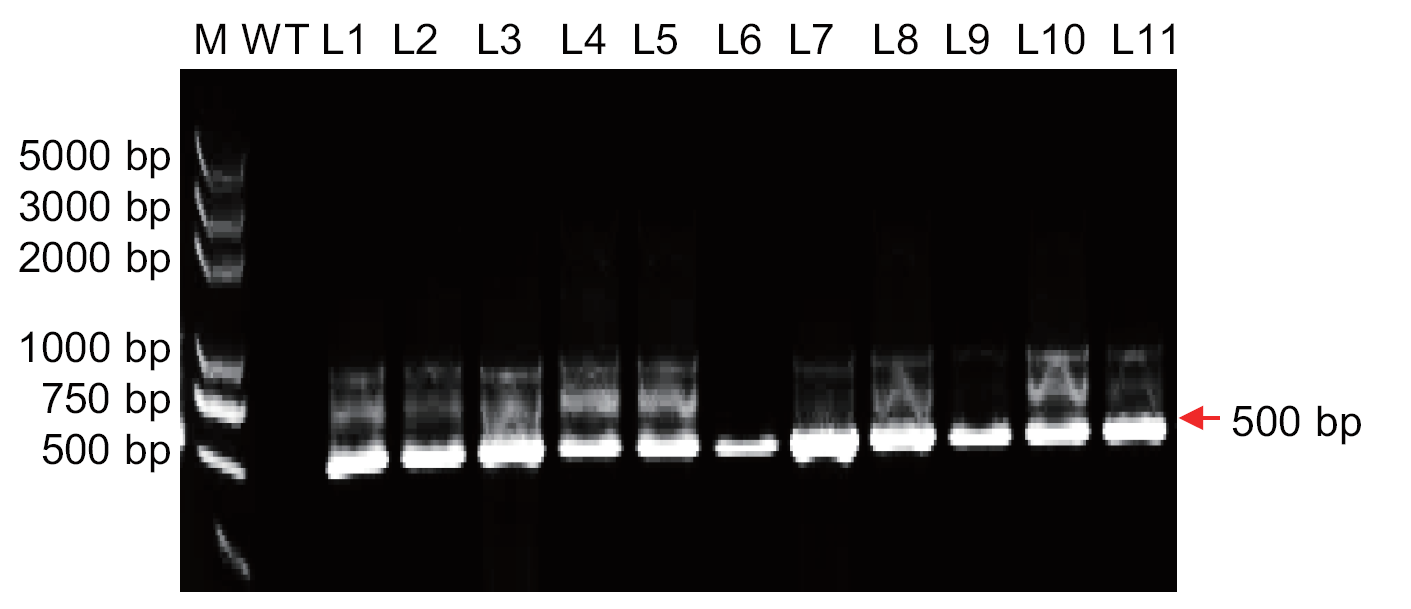
图2 PCR鉴定阳性突变体植株 M:DL 2000 DNA marker,WT:wide type,1-11:不同突变体植株。红色箭头表示目标片段(MeHPT 基因),下同
Fig. 2 PCR identification of the positive mutant plants M: DL 2000 bp DNA marker; WT:wide type; 1-11: different mutant lines. The red arrow indicates the target fragment(MeHPT gene), the same below
| Type | MeHNL-sgRNA target sites | Indel | Lines |
|---|---|---|---|
| WT | CTTGGAGAAACTCCCTCAAGGGGAAAAGGTCATCA | WT | |
| 1 | CTTGG - - - - - - - - - - - - - - - -- - - GGAAAAGGTCATCA CTTGG - - - - - - - - - - - - - - - -- - - GGAAAAGGTCATCA | -16 | L1 |
| 2 | CTTGGAGAAACTCCCTCAAGGGGAAAAAGGTCATCA CTTGGAGAAACTCCC - -AAGGAGAAGAGGTCATCA | +1, -2, 2 | L2, L7 |
| 3 | CTTGGAGAAACTCCCTCCAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCAAAAAGGGGAAAAGGTCATCA | +1, +3 | L3, L11 |
| 4 | CTTGGAGAAACTCCCTCTAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCTAAGGGGAAAAGGTCATCA | +1 | L4 |
| 5 | CTTGGAGAAACTCCCTCCAAGGGGAAAAGGTCATCA CTTGGAGAAACT- - - - - - - - - - - - - - - - - - - - GTCATCA | +1, -16 | L5 |
| 6 | CTTGGAGAAACTCCCTCAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCAAAAAGGGGAAAAGGTCATCA | +3 | L6, L9, L16 |
| 7 | CTTGGAGAAACTCCCTCGAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCC - - - AGGGGAAAAGGTCATCA | +1, -3 | L10 |
| 8 | CTTGGAGAAACTCCCTCTAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCCAAAGGGGAAAAGGTCATCA | +1, +2 | L12, L18, L31 |
| 9 | CTTGGAGAAACTCCCTC - - - - - - - - - - - - - - - - CATCA CTTGGAGAAACTCCCGTCAAGGGGAAAAGGTCATCA | -13, +1 | L13 |
| 10 | CTTGGAGAAACTCC- - - - - - -GGCAAAAGGTCATCA CTTGGAGAAACTAGCTCCAAGGGGAAAAGGTCATCA | -7, +1, +1, 2 | L14 |
| 11 | CTTGGAGAAACTCCCTC- AGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTC- AGGGGAAAAGGTCATCA | -1 | L15 |
| 12 | CTTGGAGAAACTCCCTCAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCC- -AATAAGGGGAAAAGGTCATCA | -2, +3 | L17 |
| 13 | CTTGGAGAAACTCCCTCCAAGGGGAAAAGGTCA CTTGGAGAAACTCCC- -AAAAGGGGAAAAGGTCATCA | +1, -2, +2 | L19, L32 |
| 14 | CTTGGAGAAACTCCCTCCAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCT- -TCA AAGGGGAAAAGGTCATCA | +1, -2, +3 | L21 |
| 15 | CTTGGAGAAACTCCCTCAAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCTGGAAGGGGAAAAGGTCATCA | +1, +3 | L24 |
| 16 | CTCTGAACCCTTATTGACTTTCTTGGAGAAACTCCCTCAAGGGG CTCTGAA - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -AAGGGG | -31 | L25 |
| 17 | CTTGGAGAAA - - - - - - - - AGGGGAAAAGGTCATCA CTTGGAGAAA - - - - - - - - AGGGGAAAAGGTCATCA | -8 | L26 |
| 18 | CTTGGAGAAACTCCCTCAAG - - - - - - - - - - - - - - - - - - TCATCA CTTGGAGAAA - - - - - - GGGGAAATGAAGGGGAAAAGGTCATCA | -9, -7, +9 | L29 |
表2 突变体株系中MeHNL基因靶位点突变类型
Table 2 Mutation types of MeHNL gene target sites in mutant plants
| Type | MeHNL-sgRNA target sites | Indel | Lines |
|---|---|---|---|
| WT | CTTGGAGAAACTCCCTCAAGGGGAAAAGGTCATCA | WT | |
| 1 | CTTGG - - - - - - - - - - - - - - - -- - - GGAAAAGGTCATCA CTTGG - - - - - - - - - - - - - - - -- - - GGAAAAGGTCATCA | -16 | L1 |
| 2 | CTTGGAGAAACTCCCTCAAGGGGAAAAAGGTCATCA CTTGGAGAAACTCCC - -AAGGAGAAGAGGTCATCA | +1, -2, 2 | L2, L7 |
| 3 | CTTGGAGAAACTCCCTCCAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCAAAAAGGGGAAAAGGTCATCA | +1, +3 | L3, L11 |
| 4 | CTTGGAGAAACTCCCTCTAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCTAAGGGGAAAAGGTCATCA | +1 | L4 |
| 5 | CTTGGAGAAACTCCCTCCAAGGGGAAAAGGTCATCA CTTGGAGAAACT- - - - - - - - - - - - - - - - - - - - GTCATCA | +1, -16 | L5 |
| 6 | CTTGGAGAAACTCCCTCAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCAAAAAGGGGAAAAGGTCATCA | +3 | L6, L9, L16 |
| 7 | CTTGGAGAAACTCCCTCGAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCC - - - AGGGGAAAAGGTCATCA | +1, -3 | L10 |
| 8 | CTTGGAGAAACTCCCTCTAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCCAAAGGGGAAAAGGTCATCA | +1, +2 | L12, L18, L31 |
| 9 | CTTGGAGAAACTCCCTC - - - - - - - - - - - - - - - - CATCA CTTGGAGAAACTCCCGTCAAGGGGAAAAGGTCATCA | -13, +1 | L13 |
| 10 | CTTGGAGAAACTCC- - - - - - -GGCAAAAGGTCATCA CTTGGAGAAACTAGCTCCAAGGGGAAAAGGTCATCA | -7, +1, +1, 2 | L14 |
| 11 | CTTGGAGAAACTCCCTC- AGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTC- AGGGGAAAAGGTCATCA | -1 | L15 |
| 12 | CTTGGAGAAACTCCCTCAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCC- -AATAAGGGGAAAAGGTCATCA | -2, +3 | L17 |
| 13 | CTTGGAGAAACTCCCTCCAAGGGGAAAAGGTCA CTTGGAGAAACTCCC- -AAAAGGGGAAAAGGTCATCA | +1, -2, +2 | L19, L32 |
| 14 | CTTGGAGAAACTCCCTCCAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCT- -TCA AAGGGGAAAAGGTCATCA | +1, -2, +3 | L21 |
| 15 | CTTGGAGAAACTCCCTCAAAGGGGAAAAGGTCATCA CTTGGAGAAACTCCCTCTGGAAGGGGAAAAGGTCATCA | +1, +3 | L24 |
| 16 | CTCTGAACCCTTATTGACTTTCTTGGAGAAACTCCCTCAAGGGG CTCTGAA - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -AAGGGG | -31 | L25 |
| 17 | CTTGGAGAAA - - - - - - - - AGGGGAAAAGGTCATCA CTTGGAGAAA - - - - - - - - AGGGGAAAAGGTCATCA | -8 | L26 |
| 18 | CTTGGAGAAACTCCCTCAAG - - - - - - - - - - - - - - - - - - TCATCA CTTGGAGAAA - - - - - - GGGGAAATGAAGGGGAAAAGGTCATCA | -9, -7, +9 | L29 |

图4 转基因株系MeHNL靶点区域序列比对分析 WT: Wild type,野生型。红色箭头标识突变位点
Fig. 4 Sequence alignment analysis at the target region in MeHNL transgenic lines The red arrows indicate the mutation sites

图5 野生型和突变体材料叶片中氰化物和生氰糖苷含量测定 A:生长2个月的野生型和突变体地上部表型(bars=5 cm); B:野生型和突变体材料叶片氰化物含量; C:野生型和突变体材料叶片中亚麻仁苦苷含量; D:野生型和突变体材料叶片中百脉根苷含量。* P<0.05; ** P<0.01; *** P<0.001
Fig. 5 Determination of the contents of cyanide and two raw cyanogenic glycosides in the leaves of WT and mutant plants A: The aboveground phenotypes of WT and mutant plants growth for 2 months(bars= 5 cm). B: Cyanide contents in the leaves of WT and mutant plants. C: Linamarin contents in the leaves of WT and mutant plants. D: Lotaustraline contents in the leaves of WT and mutant plants. * P<0.05; ** P<0.01; *** P<0.001
| [1] | Chaiareekitwat S, Latif S, Mahayothee B, et al. Protein composition, chlorophyll, carotenoids, and cyanide content of cassava leaves(Manihot esculenta Crantz)as influenced by cultivar, plant age, and leaf position[J]. Food Chem, 2022, 372: 131173. |
| [2] |
严华兵, 叶剑秋, 李开绵. 中国木薯育种研究进展[J]. 中国农学通报, 2015, 31(15): 63-70.
doi: 10.11924/j.issn.1000-6850.casb14110159 |
|
Yan HB, Ye JQ, Li KM. Progress of cassava breeding in China[J]. Chin Agric Sci Bull, 2015, 31(15): 63-70.
doi: 10.11924/j.issn.1000-6850.casb14110159 |
|
| [3] |
Hu W, Ji CM, Shi HT, et al. Allele-defined genome reveals biallelic differentiation during cassava evolution[J]. Mol Plant, 2021, 14(6): 851-854.
doi: 10.1016/j.molp.2021.04.009 pmid: 33866024 |
| [4] | Conn EE. Cyanogenesis - a personal perspective[J]. Acta Hortic, 1994(375): 31-44. |
| [5] | McMahon JM, White WLB, Sayre RT. REVIEW ARTICLE: Cyanogenesis in cassava(Manihot esculenta Crantz)[J]. J Exp Bot, 1995, 46(7): 731-741. |
| [6] |
Siritunga D, Sayre RT. Generation of cyanogen-free transgenic cassava[J]. Planta, 2003, 217(3): 367-373.
pmid: 14520563 |
| [7] |
Jørgensen K, Bak S, Busk PK, et al. Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology[J]. Plant Physiol, 2005, 139(1): 363-374.
pmid: 16126856 |
| [8] | Lieberman SE, Gueorguieva GA, Gill BK, et al. Transporter editing in cassava indicates local production of cyanogenic glucosides in, and export from, cassava roots[J]. Plant Biotechnol J, 2024, 22(4): 790-792. |
| [9] | Morant AV, Jørgensen K, Jørgensen C, et al. β-Glucosidases as detonators of plant chemical defense[J]. Phytochemistry, 2008, 69(9): 1795-1813. |
| [10] | Ani D, Ojila H, Abu O. Profitability of cassava processing: a case study of otukpo lga, Benue state, Nigeria[J]. Sustain Food Prod, 2019, 6: 12-23. |
| [11] | Nzwalo H, Cliff J. Konzo: from poverty, cassava, and cyanogen intake to toxico-nutritional neurological disease[J]. PLoS Negl Trop Dis, 2011, 5(6): e1051. |
| [12] | Burns A, Gleadow R, Cliff J, et al. Cassava: the drought, war and famine crop in a changing world[J]. Sustainability, 2010, 2(11): 3572-3607. |
| [13] |
Kashala-Abotnes E, Okitundu D, Mumba D, et al. Konzo: a distinct neurological disease associated with food(cassava)cyanogenic poisoning[J]. Brain Res Bull, 2019, 145: 87-91.
doi: S0361-9230(18)30324-1 pmid: 29981837 |
| [14] | Naveena K, Chinniah C, Shanthi M. Cyanogenic glycosides and plant-herbivore interactions[J]. J Entomol Zool Stud, 2021, 9(1): 1345-1350. |
| [15] |
Ceballos H, Iglesias CA, Pérez JC, et al. Cassava breeding: opportunities and challenges[J]. Plant Mol Biol, 2004, 56(4): 503-516.
doi: 10.1007/s11103-004-5010-5 pmid: 15630615 |
| [16] | Piero MN. Regeneration and RNAi-mediated downregulation of cyano-glycoside biosynthesis in cassava(Manihot esculenta, Crantz)[D]. Kenyatta University, 2014. |
| [17] | Juma BS, Mukami A, Mweu C, et al. Targeted mutagenesis of the CYP79D1 gene via CRISPR/Cas9-mediated genome editing results in lower levels of cyanide in cassava[J]. Front Plant Sci, 2022, 13: 1009860. |
| [18] | Gomez MA, Berkoff KC, Gill BK, et al. CRISPR-Cas9-mediated knockout of CYP79D1 and CYP79D2 in cassava attenuates toxic cyanogen production[J]. Front Plant Sci, 2023, 13: 1079254. |
| [19] |
Siritunga D, Arias-Garzon D, White W, et al. Over-expression of hydroxynitrile lyase in transgenic cassava roots accelerates cyanogenesis and food detoxification[J]. Plant Biotechnol J, 2004, 2(1): 37-43.
pmid: 17166141 |
| [20] | Narayanan NN, Ihemere U, Ellery C, et al. Overexpression of hydroxynitrile lyase in cassava roots elevates protein and free amino acids while reducing residual cyanogen levels[J]. PLoS One, 2011, 6(7): e21996. |
| [21] | Luo S, Ma QX, Zhong YY, et al. Editing of the starch branching enzyme gene SBE2 generates high-amylose storage roots in cassava[J]. Plant Mol Biol, 2022, 108(4/5): 429-442. |
| [22] | 李崭, 王亚杰, 陆小花, 等. 木薯MeSS III基因的CRISPR/Cas9基因编辑载体构建及验证[J]. 分子植物育种, 2020, 18(16):5367-5372. |
| Li Z, Wang YJ, Lu XH, et al. Construction and verification of CRISPR/Cas9 gene editing vector for cassava MeSSIII gene[J]. Mol Plant Breed, 2020, 18(16): 5367-5372. | |
| [23] | Bull SE, Seung D, Chanez C, et al. Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch[J]. Sci Adv, 2018, 4(9): eaat6086. |
| [24] | Wang Y, Geng M, Pan R, et al. Engineering bacterial blight-resistant plants through CRISPR/Cas9-targeted editing of the MeSWEET10a promoter in cassava[J]. bioRxiv, 2022. |
| [25] | Gomez MA, Lin ZD, Moll T, et al. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence[J]. Plant Biotechnol J, 2019, 17(2): 421-434. |
| [26] |
Zhang P, Potrykus I, Puonti-Kaerlas J. Efficient production of transgenic cassava using negative and positive selection[J]. Transgenic Res, 2000, 9(6): 405-415.
pmid: 11206969 |
| [27] |
Bolarinwa IF, Orfila C, Morgan MRA. Amygdalin content of seeds, kernels and food products commercially-available in the UK[J]. Food Chem, 2014, 152: 133-139.
doi: 10.1016/j.foodchem.2013.11.002 pmid: 24444917 |
| [28] | Du L. The biosynthesis of cyanogenic glucosides in roots of cassava[J]. Phytochemistry, 1995, 39(2): 323-326. |
| [29] | Kannangara R, Motawia MS, Hansen NKK, et al. Characterization and expression profile of two UDP-glucosyltransferases, UGT85K4 and UGT85K5, catalyzing the last step in cyanogenic glucoside biosynthesis in cassava[J]. Plant J, 2011, 68(2): 287-301. |
| [30] | Leyva-Guerrero E, Narayanan NN, Ihemere U, et al. Iron and protein biofortification of cassava: lessons learned[J]. Curr Opin Biotechnol, 2012, 23(2): 257-264. |
| [31] | McMahon Smith J, Arteca RN. Molecular control of ethylene production by cyanide in Arabidopsis thaliana[J]. Physiol Plant, 2000, 109(2): 180-187. |
| [32] |
White WLB, Arias-Garzon DI, McMahon JM, et al. Cyanogenesis in cassava. The role of hydroxynitrile lyase in root cyanide production[J]. Plant Physiol, 1998, 116(4): 1219-1225.
pmid: 9536038 |
| [33] |
Zidenga T, Siritunga D, Sayre RT. Cyanogen metabolism in cassava roots: impact on protein synthesis and root development[J]. Front Plant Sci, 2017, 8: 220.
doi: 10.3389/fpls.2017.00220 pmid: 28286506 |
| [34] | Normanly J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism[J]. Cold Spring Harb Perspect Biol, 2010, 2(1): a001594. |
| [35] |
Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development[J]. Plant Cell, 2001, 13(3): 465-480.
doi: 10.1105/tpc.13.3.465 pmid: 11251090 |
| [1] | 谭博文, 张懿, 张鹏, 王振宇, 马秋香. 木薯镁离子转运蛋白家族基因的鉴定及生物信息学分析[J]. 生物技术通报, 2024, 40(9): 20-32. |
| [2] | 赵平娟, 林晨俞, 王梦月, 张秀春, 李淑霞, 阮孟斌. 木薯SDH蛋白的序列分析及其与MeH1.2关系的研究[J]. 生物技术通报, 2024, 40(9): 74-81. |
| [3] | 侯文婷, 孙琳, 张艳军, 董合忠. 基因编辑技术在棉花种质创新和遗传改良中的应用[J]. 生物技术通报, 2024, 40(7): 68-77. |
| [4] | 隆静, 陈婧敏, 刘霄, 张一凡, 周利斌, 杜艳. 植物DNA双链断裂修复机制及其在重离子诱变和基因编辑中的作用[J]. 生物技术通报, 2024, 40(7): 55-67. |
| [5] | 周家伟, 武志强. mitoTALENs植物线粒体基因编辑载体的构建方法[J]. 生物技术通报, 2024, 40(10): 172-180. |
| [6] | 李欣格, 王美霞, 王晨阳, 马桂根, 周常勇, 王亚南, 周焕斌. 基于CRISPR/LanCas9的水稻基因编辑系统的开发和优化[J]. 生物技术通报, 2024, 40(10): 233-242. |
| [7] | 李明坤, 毕美营, 张天航, 吴翔宇, 杨培儒, 应明. UgRNA/Cas9多基因编辑法恢复根际细菌农用功能的研究[J]. 生物技术通报, 2024, 40(10): 275-287. |
| [8] | 张硕, 阚俊虎, 周家伟, 武志强. 植物线粒体基因组编辑研究进展[J]. 生物技术通报, 2024, 40(10): 41-52. |
| [9] | 杨帅朋, 屈子啸, 朱向星, 唐冬生. DNA碱基编辑技术的研究进展及在猪基因修饰中的应用[J]. 生物技术通报, 2024, 40(1): 127-144. |
| [10] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [11] | 石佳鑫, 刘凯, 朱金洁, 祁显涛, 谢传晓, 刘昌林. 基因编辑技术改良玉米株型增加杂交种产量[J]. 生物技术通报, 2023, 39(8): 62-69. |
| [12] | 肖亮, 吴正丹, 陆柳英, 施平丽, 尚小红, 曹升, 曾文丹, 严华兵. 木薯重要性状基因的研究进展[J]. 生物技术通报, 2023, 39(6): 31-48. |
| [13] | 姚晓文, 梁晓, 陈青, 伍春玲, 刘迎, 刘小强, 税军, 乔阳, 毛奕茗, 陈银华, 张银东. 二斑叶螨抗性木薯木质素合成途径基因表达特性研究[J]. 生物技术通报, 2023, 39(2): 161-171. |
| [14] | 程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291. |
| [15] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||